Einleitung
(→10.2 APSR Actor Options) |
(→6.2.4 CDA R2 Content Modules) |
||
| Zeile 1.272: | Zeile 1.272: | ||
The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, reason for anatomic pathology procedure, clinical laboratory data, specimen collection procedure including target site, performer, specimen type, specimen(s) clinical description, and tumor site in case of a cancer. | The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, reason for anatomic pathology procedure, clinical laboratory data, specimen collection procedure including target site, performer, specimen type, specimen(s) clinical description, and tumor site in case of a cancer. | ||
| + | {{:2.16.840.1.113883.2.6.60.6.10.5}} | ||
| − | |||
====6.2.4.2 Intraoperative Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.2==== | ====6.2.4.2 Intraoperative Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.2==== | ||
| Zeile 1.315: | Zeile 1.315: | ||
The Report Textual Summary section is an optional sub-section of the Diagnostic Conclusion section. This section contains a textual summary of the AP report, which can be extracted from the document and inserted into other clinical documents. It addresses the use case where authors of other medical documents feel the need to include a segment such as "...the pathology report states '[...]'", the text content of this section filling the brackets. | The Report Textual Summary section is an optional sub-section of the Diagnostic Conclusion section. This section contains a textual summary of the AP report, which can be extracted from the document and inserted into other clinical documents. It addresses the use case where authors of other medical documents feel the need to include a segment such as "...the pathology report states '[...]'", the text content of this section filling the brackets. | ||
| − | |||
===6.2.5 CDA R2 <entry> Content Modules=== | ===6.2.5 CDA R2 <entry> Content Modules=== | ||
Version vom 20. Juni 2016, 18:22 Uhr
Dieses Material ist Teil des Leitfadens Pathologiebefund.
|
Foreword
This is a supplement to the IHE Pathology and Laboratory Medicine Technical Framework V7.0. Each supplement undergoes a process of public comment and trial implementation before being incorporated into the volumes of the Technical Frameworks.
This supplement is published on <Month XX, 2016> for Public Comment. Comments are invited and may be submitted at http://www.ihe.net/<domain>/<domain>comments.cfm. In order to be considered in development of the Trial Implementation version of the supplement, comments must be received by <Month XX, 201X>.
This supplement describes changes to the existing technical framework documents.
“Boxed” instructions like the sample below indicate to the Volume Editor how to integrate the relevant section(s) into the relevant Technical Framework volume.
Replace Section X.X by the following:
Where the amendment adds text, make the added text bold underline. Where the amendment removes text, make the removed text bold strikethrough. When entire new sections are added, introduce with editor’s instructions to “add new text” or similar, which for readability are not bolded or underlined.
General information about IHE can be found at: http://www.ihe.net.
Information about the IHE Pathology and Laboratory Medicine domain can be found at: http://ihe.net/IHE_Domains.
Information about the structure of IHE Technical Frameworks and Supplements can be found at: http://ihe.net/IHE_Process and http://ihe.net/Profiles.
The current version of the IHE Technical Framework (if applicable) can be found at: http://ihe.net/Technical_Frameworks.
<Comments may be submitted on IHE Technical Framework templates any time at http://ihe.net/ihetemplates.cfm. Please enter comments/issues as soon as they are found. Do not wait until a future review cycle is announced.
Introduction to this Supplement
This supplement complements volume 1 of the PaLM technical framework with the description of the APSR 2.0 content profile, and volume 3 with the bindings, content modules and value sets, which specify this profile.
Open Issues and Questions
None yet
Closed Issues
APSR-07 – Representing the hierarchy of specimens in an entry : This APSR supplement enables to represent the hierarchy of specimens at the CDA level 3. The operations on specimen and production of child specimens are tracked in the “Procedure Steps” section, which has a level 3 entry.
APSR-10 – Observation related to multiple specimens: For example tumor staging may require combining data from multiple specimens (e.g., a breast excision with positive margins followed by a re-excision with clear margins – in this case the tumor size may be a composite of measurements from both specimens. Another example is staging of ovarian carcinomas with multiple biopsies of pelvis, peritoneum, nodes, omentum, etc.). To accommodate these use cases, the specimen organizer is able to represent either a single specimen or a group of specimens investigated together.
APSR-11 – Derivative specimens:Specimens derived from primary specimens for ancillary studies, which may be sent to a reference lab or done in another part of the same institution, are included in the scope of this profile. The results produced on a derived specimen are attached to this specimen in the report.
VOLUME 1 -PROFILES
10 Anatomic Pathology Structured Report (APSR) Profile
This content profile describes an anatomic pathology structured report (APSR) as a digital document to be shared or exchanged between pathology laboratories and other care providers and institutions.
Anatomic pathology structured reports document the findings on specimens removed from patients for diagnostic or therapeutic reasons. This information can be used for patient care, clinical research and epidemiology. Standardizing and computerizing anatomic pathology reports is necessary to improve the quality of reporting and to facilitate the exchange the exchange and reuse of the content of these reports.
This content profile describes a digital anatomic pathology report shared in a human-readable format, which may include images, and which also contains findings and observations in a machine-readable format, to facilitate the integration of these into the database of a consumer system, and to enable the application of automated reasoning to this content.
The scope of this IHE content profile covers all fields of anatomic pathology (cancers, benign neoplasms as well as non-neoplastic conditions) as well as cytopathology.
Goldsmith, J.D., et al., “Reporting guidelines for clinical laboratory reports in surgical pathology” Arch Pathol Lab Med, 2008. 132(10): p. 1608-16, is the first source of specification for this content profile. This article delineates the required, preferred, and optional elements which should be included in any report of surgical pathology.
This source is complemented by the “cancer checklists” produced by the College of American Pathologists, and by the “comptes rendus d’anatomopathologie : données minimales à renseigner pour une tumeur primitive” produced by the French society of pathology (SFP [1]) for the French cancer institute (INCa [www.e-cancer.fr]).
This profile has also benefited from the guidance on cancer AP reports provided by the North-American Association of Central Cancer Registries; some of the example snippets captured in the profile leverage the NAACCR Standards for Cancer Registries, Volume V, Pathology Laboratory Electronic Reporting.
10.1 APSR Actors/Transactions
This section defines the actors, transactions, and/or content modules in this profile. General definitions of actors are given in the Technical Frameworks General Introduction Appendix A published here.
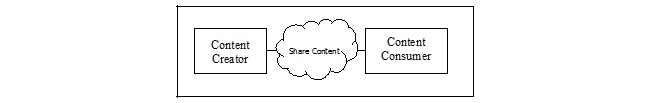
Figure 10.1-1 shows the actors directly involved in the APSR Profile and the direction that the content is exchanged.
A product implementation using this profile must group actors from this profile with actors from a workflow or transport profile to be functional. The grouping of the content module described in this profile to specific actors is described in more detail in the “Required Actor Groupings” section below.
Figure 10.1-1 APSR Actor Diagram
Table 10.1-1 lists the content module(s) defined in the APSR profile. To claim support with this profile, an actor shall support all required content modules (labeled “R”) and may support optional content modules (labeled “O”).
Table 10.1-1: <Profile Acronym>Profile - Actors and Content Modules
| Actors | Content Modules | Optionality | Reference |
|---|---|---|---|
| Content
Creator |
Anatomic Pathology Structured Report 1.3.6.1.4.1.19376.1.8.1.1.1 |
R | PaLM TF-3: 6.3.1.2 |
| Content
Consumer |
Anatomic Pathology Structured Report 1.3.6.1.4.1.19376.1.8.1.1.1 |
R | PaLM TF-3: 6.3.1.2 |
10.1.1 Actor Descriptions and Actor Profile Requirements
Most requirements are documented in Content Modules (Volume 3). This section documents any additional requirements on profile’s actors.
10.2 APSR Actor Options
Options that may be selected for each actor in this profile are listed in the table 10.2-1. These options are further detailed in PCC Technical Framework Volume 2 as indicated in the rightmost column.
Table 10.2-1 Anatomic Pathology Structured Report - Actors and Options
| Actor | Option Name | Reference |
|---|---|---|
| Content
Creator |
None | |
| Content
Consumer |
View Option (1)
Document Import Option (1) Section Import Option (1) |
PCC TF-2:3.1.1
PCC TF-2:3.1.2 PCC TF-2:3.1.3 |
Note 1: The Content Consumer Actor shall support at least one of these options.
10.3 APSR Required Actor Groupings
An Actor from this profile (Column 1) shall implement all of the required transactions and/or content modules in this profile in addition to all of the transactions required for the grouped actor (Column 2).
In some cases, required groupings are defined as at least one of an enumerated set of possible actors; this is designated by merging column one into a single cell spanning multiple potential grouped actors. Notes are used to highlight this situation.
Section 10.5 describes some optional groupings that may be of interest for security considerations and section 10.6 describes some optional groupings in other related profiles.
Table 10.3-1: Anatomic Pathology Structured Report - Required Actor Groupings
| XD-LAB Actor | Actor to be grouped with | Reference | Content Bindings Reference |
|---|---|---|---|
| Content
Creator |
ITI XDS.b Document Source OR ITI XDM Portable Media Creator OR ITI XDR Document Source |
ITI TF-1:10
|
|
| Content
Consumer |
ITI XDS.b Document Consumer OR ITI XDM Portable Media Consumer OR ITI XDR Document Recipient |
ITI TF-1:10
ITI TF-1:16
ITI TF-1:15 |
Note 1: Each actor of APSR SHALL be grouped with at least one of the ITI actors listed in its table row.
10.4 APSR Overview
10.4.1 Concepts
This content profile represents a common digital document model applicable to any structured report for surgical pathology in all fields of anatomic pathology (cancers, benign neoplasms, non-neoplastic conditions) as well as for cytopathology.
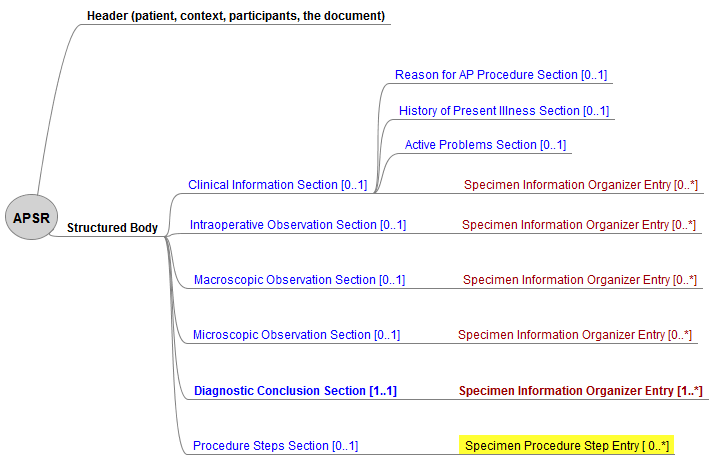
This common model is composed of a header conveying the context of care (patient, care providers, pathologists, laboratories, order, act documented …) and a body. The body organizes the human-readable content of the report in a number of sections. Each section may also provide machine-readable content in an “entry” embedded in the section. This common model defines the order of appearance, cardinalities and internal structure of each section, and of each entry embedded in each section.
Figure 10.4.1-1 shows this general model applicable to any pathology digital report.
Figure 10.4.1-1 Common model for a digital anatomic pathology report
Note 1: The only section that is mandatory is the Diagnostic Conclusion section.
4.1.2 Document Content Modules
4.1.2.1 Anatomic Pathology Structured Report (APSR)
This document content module represents the generic set of constraints applied to any structured report for surgical pathology in all fields of anatomic pathology (cancers, benign neoplasms as well as non-neoplastic conditions) as well as for Cytopathology.
This document content module is identified by templateId 1.3.6.1.4.1.19376.1.8.1.1.1
The body of this document content module has the hierarchy of sections and entries depicted by figure 4.1.2.1-1. The only mandatory section is the Diagnostic Conclusion Section. And the only mandatory entry is the Specimen Information Organizer Entry below this section.
Figure 4.1.2.1-1 Hierarchy of the body for APSR document content module
4.2 APSR Options
Table 4.2-1 summarizes the options that actors may take for this content profile. Dependencies between options when applicable are specified in notes. These options are summarized below the table, and further detailed in PCC TF-2, as indicated in the right column of the table.
Table 4.2-1 Actors and Options
| Actor | Options | Domain, Vol & Section |
|---|---|---|
| Content
Consumer |
View Option (1) | PCC TF-2:3.1.1 |
| Content
Consumer |
Document Import Option (1) | PCC TF-2:3.1.2 |
| Content
Consumer |
Section Import Option (1) | PCC TF-2:3.1.3 |
Note 1: The Actor shall support at least one of these options.
4.3 APSR Actor Groupings and Profile Interactions
It is expected that the sharing of anatomic pathology structured reports will occur in an environment where the physician offices and hospitals have a coordinated infrastructure that serves the information sharing needs of this community of care. Several mechanisms are supported by IHE profiles:
- A registry/repository-based infrastructure is defined by the IHE Cross-Enterprise Document Sharing (XDS) and other IHE Integration Profiles such as patient identification (PIX & PDQ), and notification of availability of documents (NAV).
- A media-based infrastructure is defined by the IHE Cross-Enterprise Document Media Interchange (XDM) profile.
- A reliable messaging-based infrastructure is defined by the IHE Cross-Enterprise Document Reliable Interchange (XDR) profile.
- All of these infrastructures support Security and privacy through the use of the Consistent Time (CT) and Audit Trail and Node Authentication (ATNA) profiles.
For more details on these profiles, see the IHE IT Infrastructure Technical Framework
Such an infrastructure is assumed by the use cases described in this Profile.
A content binding describes how the payloads used in IHE transactions are related to and/or constrained by the data elements contained within the content sent or received in those transactions. The APSR Content Profile applies one binding, which is used when grouping the Content Creator with the IHE ITI XDS, XDM or XDR Integration Profiles.
The content and the binding are described in Volume 3 of the IHE Anatomic Pathology Technical Framework.
4.4 APSR Process Flow
4.4.1 Use Cases
4.4.1.1 Use case 1: General case
Barbara Breast visits Sammy Surgeon for removal of a breast tumor. Sammy Surgeon orders the Requested Procedure “Breast surgical specimen - pathological examination” and sends the specimen(s) to the anatomic pathology department.
Specimen(s) is (are) accessioned by the anatomic pathology department. The staff performs a macroscopic examination of the specimen(s); gross imaging is performed if needed. The specimen(s) are processed for microscopic examination and other special ancillary techniques or tissue banking if needed. During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process, pathologist queries the Content Creator application for the appropriate APSR template, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs the document.
4.4.1.2 Use case 2: Specimen collector is not the ordering physician
Patricia Pathologist collects bone marrow from Peter Patient in the clinical ward.
Specimen(s) is (are) accessioned by the anatomic pathology department. The staff performs a macroscopic examination of the specimen(s); gross imaging is performed if needed. The specimen(s) are processed for microscopic examination and other special ancillary techniques or tissue banking if needed. During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process, pathologist queries the Content Creator for the appropriate APSR template, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs the document.
4.4.1.3 Use case 3: Multi-step reporting
Barbara Breast visits Sammy Surgeon for removal of a breast tumor. Sammy Surgeon orders the Requested Procedure “Breast surgical specimen - Frozen sections & pathological examination” and sends the specimen(s) to the anatomic pathology department.
Specimen(s) is (are) accessioned by the anatomic pathology department. The staff performs a macroscopic examination of the specimen(s), gross imaging is performed if needed. The specimen(s) are processed for intraoperative observation if needed, and tissue banking if needed (e.g., for research purpose). During the imaging interpretation process of frozen sections, microscopic imaging is performed if needed. At the end of the interpretation process, pathologist queries the Content Creator for the appropriate APSR template, fills the intraoperative observation section, binds some relevant images and/or regions of interest to specific observation(s) if needed, validates and signs (i.e., legally authenticates) the preliminary APSR.
The day after, the specimen(s) are processed for microscopic examination and other special ancillary techniques if needed. During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process, pathologist queries the Content Creator for the preliminary APSR, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs (i.e., legally authenticates) the final APSR.
4.4.1.4 Use case 4: Opinion request
There are various situations in which an opinion request is issued: External expert consultation (requested by Philip Pathologist, often before a final report is issued). Second opinion request (usually made by a treating physician or patient/family, or lawyer/court in malpractice cases). External slide review (usually routine review of slides required by protocols in an outside treating institution). These may vary in terms or workflow and even the materials accessed by the outside lab.
Requiring pathologist Philip Pathologist asks for second opinion for the case of Peter Patient diagnosed as lymphoma. He sends block(s) or slide(s) or shares/sends whole slide images to Patricia pathologist, requesting her expertise on this material. He uses the Content Creator application to derive the anatomic pathology opinion request document from the preliminary APSR of Peter Patient.
Philip Pathologist later on uses his Content Consumer application to view and import the APSR from Patricia Pathologist. He uses this report to finalize and issue his own APSR in his application acting as a Content Creator.
Requested pathologist
Patricia Pathologist accepts to deliver second opinion about the case of Peter Patient diagnosed as lymphoma.
Block(s)
The specimen(s) are processed for microscopic examination and other special ancillary techniques if needed. During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process, Patricia Pathologist queries the Content Creator for the appropriate APSR template, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs the document.
Slide(s)
During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process, Patricia Pathologist queries the Content Creator for the appropriate APSR template, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs the document.
Whole slide image(s)
At the end of the interpretation process, Patricia Pathologist queries the Content Creator for the appropriate APSR template, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs the document.
4.5 APSR Security considerations
4.5.1Integrity
The choice on whether the digital signature is performed at the transaction level (XDS, XDM, XDR) or at the content level is left up to national extensions. If the report is digitally signed, the person having signed it SHALL be the person represented in the legalAuthenticator element of the CDA header.
4.5.2 Confidentiality
In the context of patient care coordination the anatomic pathology report described in this profile contains patient personal data, and as such must be handled in conformance to the local privacy policies.
In other contexts such as public health, surveillance, research, appropriate content anonymization and patient identification pseudonymization may be required by local policies.
4.5.3 Auditability
Addressed by the CT and ATNA profiles from ITI TF.
Volume 3 – Content Modules
1 Introduction
Insert the text from the same section in volume 1 of the PAT TF
1.1 Overview of the Anatomic Pathology Technical Framework
Insert the text from the same section in volume 1 of the PAT TF
1.2 Overview of Volume 3
The IHE Technical Framework is based on actors that interact through transactions using some form of content.
Actors are
information systems or components of information systems that produce, manage,
or act on information associated with operational activities in the enterprise.
Transactions are
interactions between actors that transfer the required information through
standards-based messages.
Content profiles
specify how the payload of a transaction fits into a specific use of that
transaction. A content profile has three main parts. The first part describes
the use case. The second part is binding to a specific IHE transaction, which
describes how the content affects the transaction. The third part is a Content
Module, which describes the payload of the transaction. A content module is
specified so as to be independent of the transaction in which it appears. This
overall content module is itself an assemblage of smaller content modules,
which in turn may assemble smaller content modules, conforming to the chosen
standard.
In particular,
the Anatomic Pathology Technical Framework provides a set of content profiles
for the sharing of persistent clinical document produced by the anatomic
pathology domain.
This Volume 3
specifies the content modules produced at various granularity levels (from a
whole clinical document to a tiny reusable piece of coded data) by the Anatomic
Pathology domain of IHE for its own content profiles.
Some of these
content modules produced here, may be used by content modules of higher
granularity from other domains (e.g., Patient Care Coordination).
Some of these
content modules produced here, may leverage content modules of lower
granularity from other domains (e.g., PCC, RAD, etc.).
1.3 Audience
Insert the text from the same section in volume 1 of the PAT TF
1.4 Relationship to Standards
Insert a simplified version of the text from the same section in volume 1 of the PAT TF
1.5 Relationship to Real World Architecture
Insert the text from the same section in volume 1 of the PAT TF
1.6 Conventions
Insert a simplified version of the text from the same section in volume 1 of the PAT TF
1.7 Scope Introduced in the Current Year
Content Modules for the APSR Profile
1.8 Copyright Permission
Health Level Seven, Inc. has granted permission to the IHE to reproduce tables from the HL7 standard. The HL7 tables in this document are copyrighted by Health Level Seven, Inc. All rights reserved. Material drawn from these documents is credited where used.
1.9 Glossary
The glossary of the Anatomic Pathology Technical Framework is centralized in PAT TF-1:1.12.
2 Content Modules – Basic Principles
This Volume 3 of the PAT TF organizes content modules categorically by the base standard. At present, PAT TF-3 uses only one base standard, CDA Release 2.0, but this is expected to change over time. Underneath each standard, the content modules are organized using a very coarse hierarchy inherent to the standard.
Each content module can be viewed as the definition of a "class" in software design terms, and has associated with it a name. Like "class" definitions in software design, a content module is a "contract", and the PAT TF-3 defines that contract in terms of constraints that must be obeyed by instances of that content module. Each content module has a name, also known as its template identifier. The template identifiers are used to identify the contract agreed to by the content module. The Anatomic Pathology Technical Committee is responsible for assigning the template identifiers to each content module.
Like classes, content modules may inherit features of other content modules of the same type (e.g., Document, Section or Entry) by defining the parent content module that they inherit from. They may not inherit features from a different type.
Constraints that apply to any content module will always apply to any content modules that inherit from it. Thus, the "contracts" are always valid down the inheritance hierarchy.
The PAT TF-3 uses the convention that a content module cannot have more than one parent (although it may have several ancestors). This convention is not due to any specific technical limitation of the technical framework, but does make it easier for software developers to implement content modules.
Each content module has a list of data elements that are required (R), required if known (R2), conditional (C) or optional (O).
Other data elements may be included in an instance of a content module over what is defined by the PAT TF-3. Content consumers are not required to process these elements, and if they do not understand them, must ignore them. Thus, it is not an error to include more than is asked for, but it is an error to reject a content module because it contains more than is defined by the framework. This allows value to be added to the content modules delivered internationally in this framework, through national extensions built by the national IHE organizations in various countries. It further allows content modules to be defined later by IHE that are refinements or improvements over previous content modules.
3 IHE Transactions
This section defines each IHE transaction in detail, specifying the standards used, and the information transferred.
3.1 Cross Enterprise Document Content Transactions
At present, all transactions used by the Anatomic Pathology Content Profiles appear in ITI TF-2a and ITI TF-2b.
General Options defined in content profiles for a Content Consumer are listed below.
3.1.1 View Option
A Content Consumer that supports the View Option shall be able to:
1. Use the appropriate XD* transactions to obtain the document along with associated necessary metadata.
2. Render the document for viewing. This rendering shall meet the requirements defined for CDA Release 2 content presentation semantics (See Section 1.2.4 of the CDA Specification: Human readability and rendering CDA Documents). CDA Header information providing context critical information shall also be rendered in a human readable manner. This includes at a minimum the ability to render the document with the stylesheet specifications provided by the document source, if the document source provides a stylesheet. Content Consumers may optionally view the document with their own stylesheet, but must provide a mechanism to view using the source stylesheet.
3. Support traversal of links for documents that contain links to other documents managed within the sharing framework.
4. Print the document to paper.
3.1.2 Document Import Option
This Option requires that the View Option be supported. In addition, the Content Consumer that supports the Document Import Option shall be able to support the storage of the entire APSR document (as provided by the sharing framework, along with sufficient metadata to ensure its later viewing). The machine-readable content (from the entry elements) shall also be imported. This Option requires the proper tracking of the document origin. Once a document has been imported, the Content Consumer shall offer a means to view the document without the need to retrieve it again from the sharing framework. When the document is used after it was imported, a Content Consumer may choose to access the sharing framework to find out if the related Document viewed has been deprecated or replaced.
3.1.3 Section Import Option
This Option requires that the View Option be supported. In addition, the Content Consumer that supports the Section Import Option shall be able to support the import of one or more sections of the APSR document (along with sufficient metadata to link the data to its source). The machine-readable content (from the entry elements beneath the imported sections) shall also be imported. This Option requires the proper tracking of the document section origin. Once sections have been selected, a Content Consumer shall offer a means to copy the imported section(s) into local data structures. When a section is used after it is imported, a Content Consumer may choose to access the sharing framework to find out if the related information has been updated.
4 IHE Anatomic Pathology Bindings
This section describes how the payload used in a transaction of an IHE profile is related to and/or constrains the data elements sent or received in those transactions. This section is where any specific dependencies between the content and transaction are defined.
A content profile can define multiple bindings. Each binding should identify the transactions and content to which it applies.
The source for all required and optional attributes have been defined in the bindings below. Three tables describe the three main XDS object types: XDSDocumentEntry, XDSSubmissionSet, and XDSFolder. XDSSubmissionSet and XDSDocumentEntry are required. Use of XDSFolder is optional. These concepts are universal to XDS, XDR and XDM.
The structure of these three tables is presented in PCC TF-2:4
4.1 Anatomic Pathology Document Binding to XDS, XDM and XDR
This binding defines a transformation that generates metadata for the XDSDocumentEntry element of appropriate transactions from the XDS, XDM and XDR profiles given a medical document and information from other sources. The medical document refers to the document being stored in a repository that will be referenced in the registry. The other sources of information include the configuration of the Document Source actor, the Affinity Domain, the site or facility, local agreements, other documents in the registry/repository, and this content profile.
In many cases, the CDA document is created for the purposes of sharing within an affinity domain. In these cases the context of the CDA and the context of the affinity domain are the same, in which case the following mappings shall apply.
In other cases, the CDA document may have been created for internal use, and are subsequently being shared. In these cases the context of the CDA document would not necessarily coincide with that of the affinity domain, and the mappings below would not necessarily apply.
4.1.1XDS DocumentEntry Metadata
The general table describing the XDSDocumentEntry Metadata requirements for IHE domains is shown in PCC TF-2:4.1.1
The sub-sections below list the only requirements which are specific to the Anatomic Pathology Domain, and which supersede those from the general table mentioned above.
4.1.1.1XDS DocumentEntry.formatCode
The values of formatCode per document template are listed in table 5.6-1.
The associated codingScheme Slot SHALL be 1.3.6.1.4.1.19376.1.2.3 in all cases.
4.1.1.2XDS DocumentEntry.eventCodeList
This metadata provides a means to index anatomic pathology reports by reportable conditions (e.g., certain types of tumors…) so as to facilitate later queries in a registry of shared clinical documents. The conclusions coded in the entry element of the Diagnostic Conclusion section are good candidates for this metadata.
4.1.1.3XDS DocumentEntry.parentDocumentRelationship
The Anatomic Pathology document Content Modules only permit the “replace” relationship between instances of APSR documents.
Thus, XDSDocumentEntry.parentDocumentRelationship is constrained to the "RPLC" (replace) value. The new document issued replaces completely the parent one, which will be considered as deprecated.
4.1.2XDS SubmissionSet Metadata
The submission set metadata is as defined for XDS, and is not necessarily affected by the content of the clinical document. Metadata values in an XDSSubmissionSet with names identical to those in the XDSDocumentEntry may be inherited from XDSDocumentEntry metadata, but this is left to affinity domain policy and/or application configuration.
This content format uses the submission set to create a package of information to send from one provider to another. All documents or images referenced by the Anatomic Pathology Structured Report in this Package must be present (at least as references in the case of images) in the submission set.
4.1.3XDS Folder Metadata
No specific requirements identified.
4.1.4 Configuration
The Anatomic Pathology Content Profiles using this binding require that Content Creators and Content Consumers be configurable with institution and other specific attributes or parameters. Implementers should be aware of these requirements to make such attributes easily configurable.
5 Namespaces and Vocabularies
5.1 OID tree of PAT TF
1.3.6.1.4.1.19376.1.81.3.6.1.4.1.19376.1.8 is the OID of the IHE Anatomic Pathology domain:
All exchangeable objects specified by this domain are identified by OIDs built on this root:
Branch 1.3.6.1.4.1.19376.1.8.1 is dedicated to CDA Content Modules created by this domain
1.3.6.1.4.1.19376.1.8.1.1 is the OID of the generic Document Content Module
Sub-branch 1.3.6.1.4.1.19376.1.8.1.2 is dedicated to Section Content Modules
Sub-branch 1.3.6.1.4.1.19376.1.8.1.4 is dedicated to Element Content Modules
1.3.6.1.4.1.19376.1.8.1.4.4 is the OID of the Specimen Information Organizer
1.3.6.1.4.1.19376.1.8.1.4.8 is the OID of the Problem Organizer
1.3.6.1.4.1.19376.1.8.1.4.9 is the OID of the generic anatomic pathology (AP) observation template
1.3.6.1.4.1.19376.1.8.1.4.10 is the OID of the Procedure Steps template
Branch 1.3.6.1.4.1.19376.1.8.2 is dedicated to terminologies defined by this domain
Sub-branch 1.3.6.1.4.1.19376.1.8.2.1 is dedicated to PathLex
Branch 1.3.6.1.4.1.19376.1.8.5 is dedicated to Value Sets defined by this domain.
Branch 1.3.6.1.4.1.19376.1.8.9 is used to identify instances in the examples built by the PAT TF.
Notes on other IHE OIDs used in the AP domain:
1.3.6.1.4.1.19376.1.3.1.2 is the OID of the Specimen Collection Procedure template
5.2 Terminologies and controlled coded vocabularies
This section lists the terminologies and the coded vocabularies referenced by this Volume 3 of the IHE PAT TF.
Table 5.2-1 Anatomic Pathology Terminologies and Coded Vocabularies
 |
Could this be contributed by ArtDecor??? GH |
| codeSystem | codeSystemName | Description | Owner |
|---|---|---|---|
| 2.16.840.1.113883.6.1 | LOINC | Logical Observation Identifier Names and Codes | Regenstrief Institute |
| 2.16.840.1.113883.6.96 | SNOMED-CT | Systematized Nomenclature of Medicine – Clinical Terms | IHTSDO |
| 1.3.6.1.4.1.19376.1.5.3.2 | IHEActCode | Vocabulary defined by IHE PCC in PCC TF-2:5.1.2 | IHE PCC |
| 2.16.840.1.113883.6.3 | ICD-10 | International Classification of Diseases revision 10 | WHO |
| 2.16.840.1.113883.6.43 | ICD-O-3 | International Classification of Diseases for Oncology revision 3 | WHO |
| PubCan | A Public Database of Human Cancers http://www.pubcan.org | WHO | |
| 1.2.250.1.213.2.11 | ADICAP Thesaurus | French thesaurus of lesions in anatomic pathology | ADICAP |
| 1.2.250.1.213.2.12 | SNOMED International (3.5) | Systematized Nomenclature of Medicine | ASIP santé
|
| 1.3.6.1.4.1.19376.1.8.2.1 | PathLex | Temporary terminology covering the scope of anatomic pathology observation results and specimen collection procedure code | IHE-PAT |
| 2.16.840.1.113883.15.6 | TNM 7th edition | Internationally agreed-upon standards to describe and categorize cancer stages and progression http://www.uicc.org/resources/tnm | Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC) |
| 2.16.840.1.113883.15.7 | TNM 6th edition | Internationally agreed-upon standards to describe and categorize cancer stages and progression http://www.uicc.org/resources/tnm | Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC) |
| 2.16.840.1.113883.15.8 | TNM 5th edition | Internationally agreed-upon standards to describe and categorize cancer stages and progression http://www.uicc.org/resources/tnm | Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC) |
| 1.2.276.0.76.3.1.131.1.5.1 | DKG Coding Scheme | Internationally agreed-upon standards to describe and categorize cancer stages and progression, adapted for Germany | DKG (Deutsche Krebsgesellschaft) |
5.3 Value Sets
The value sets defined or referenced by this Volume 3 of the IHE PAT TF are listed in the separate appendix spreadsheet “IHE_PAT_Suppl_APSR_AppendixValue_Sets”.
5.4 Namespaces
5.4.1 Namespace protecting extensions to the CDA schema
There is currently one single extension to the CDA.xsd schema used in PAT TF-3. This extension has been created by IHE LAB and is protected by this particular namespace in document instances: xmlns:lab="urn:oid:1.3.6.1.4.1.19376.1.3.2"
5.5 References to Content Modules built outside of IHE PAT TF
The Content Modules specified in this Volume 3 of the PAT TF leverage a number of Content Modules (currently CDA templates) produced and maintained by other groups, including other domains of IHE. Table 5.5-1 lists them.
Table 5.5-1 External Content Modules referenced by PAT TF-3
| templateId | Standard | Definition | Source of Specification |
|---|---|---|---|
| 1.3.6.1.4.1.19376.1.5.3.1.3.1 | CDA R2 | Reason for referral | IHE PCC TF-2:6.3.3.1.2 |
| 1.3.6.1.4.1.19376.1.5.3.1.3.4 | CDA R2 | History of present illness | IHE PCC TF-2:6.3.3.2.1 |
| 1.3.6.1.4.1.19376.1.5.3.1.3.6 | CDA R2 | Active Problems | IHE PCC TF-2:6.3.3.2.3 |
| 1.3.6.1.4.1.19376.1.5.3.1.4.2 | CDA R2 | Annotation Comment | IHE PCC TF-2:6.3.4.6 |
| 1.3.6.1.4.1.19376.1.3.3.1.7 | CDA R2 | Performing laboratory | IHE LAB TF-3:2.3.3.22 |
| 1.3.6.1.4.1.19376.1.3.3.1.6 | CDA R2 | Ordering Provider (ordering physician) | IHE LAB TF-3:2.3.3.19 |
| 1.3.6.1.4.1.19376.1.3.3.1.4 | CDA R2 | Intended Recipient | IHE LAB TF-3:2.3.3.16 |
| 1.3.6.1.4.1.19376.1.3.1.6 | CDA R2 | Laboratory Observation | IHE LAB TF-3:2.3.5.11
|
| 1.3.6.1.4.1.19376.1.3.1.2 | CDA R2 | Specimen Collection Procedure | IHE LAB TF-3:2.3.5.5 (specification captured in this APSR supplement for easier readability) |
5.6 IHE Codes for Anatomic Pathology Document Templates
Any AP structured report SHALL conform to the APSR generic Content Module Identified by templateId “1.3.6.1.4.1.19376.1.8.1.1.1” , and SHALL be associated with the following metadata:
- typeCode = “11526-1”, which is the LOINC code for “Pathology study”.
- formatCode = “urn:ihe:pat:apsr:all:2010”, with associated codingScheme = “1.3.6.1.4.1.19376.1.2.3” as assigned by the ITI Domain for codes used for the purposes of cross-enterprise document sharing (XDS).
- The media type SHALL be “text/xml”.
6 Anatomic Pathology Content Modules
6.1 Conventions
In all Content Modules specified in this section, the abbreviation “AP” stands for “Anatomic Pathology”.
Various tables used in this section will further constrain the content. Within this volume, the following conventions are used:
R
A "Required" data element is one that shall always be provided. If there is information available, the data element must be present. If there is no information available, or it cannot be transmitted, the data element must contain a value indicating the reason for omission of the data.
R2
A "Required if data present" data element is one that shall be provided when a value exists. If the information cannot be transmitted, the data element shall contain a value indicating the reason for omission of the data. If no such information is available to the content creator or if such information is not available in a well identified manner (e.g., buried in a free form narrative that contains additional information relevant to other sections) or if the content creator requires that information be absent, the R2 section shall be entirely absent. The Content Creator application must be able to demonstrate that it can populate all required if known elements, unless it does not in fact gather that data. This “R2” code is the equivalent of the HL7 standard “RE” usage code. The value “R2” has been chosen in harmonization with the IHE PCC TF, which is the source of a large number of CDA R2 content modules.
O
An "Optional" data element is one that may be provided, irrespective of whether the information is available or not. If the implementation elects to support this optional section, then its support shall meet the requirement set forth for the "Required if data present" or R2.
C
A "Conditional" data element is one that is required, required if known or optional depending upon other conditions. These will have further notes explaining when the data element is required, et cetera.
6.2 HL7 CDA R2 Content Modules
6.2.1 Organization
6.2.1.1 Various Types of Content Modules
For the CDA Release 2.0 standard, the content modules are organized by:
- document: The template for a whole document.
- section: The template for a <section> element.
- entry: The template for a <entry> element.
- child element: An element of the CDA header or an element of a <section>, or an element nested at various depths below an <entry>, or an element appearing at some combination of these locations.
6.2.1.2 General constraints added by IHE PAT to a CDA R2 document
In the structured body of a CDA R2 document, a section has a narrative block (the text element), which presents the human-readable content of the section, and MAY have one entry or more. Sections MAY be nested within one another.
The content modules designed by the PAT TF bring or highlight the following constraints:
- When a section has a text element and one or more entry element, the content coded for machine-processing in the entries SHALL be completely transcribed into human-readable content in the text element.
- Conversely the text element MAY contain pieces of information, which are not available in machine-readable format in any entry element of the section.
- For a document of the Anatomic Pathology domain, the entry elements are instantiated per specimen or per group of specimens observed together. One entry contains in machine-readable format observations of the section related to the same specimen or group of specimens. Beneath an entry, the observations are organized per problem.
- The text element of the section is supposed to be also laid out per specimen or group of specimens and per problem observed.
- The APSR Content Profile leaves the layout of the text element up to the Content Creator applications, or to further constraints brought by national extensions of this profile. However, given that the text element is usually composed of free text (e.g. dictated text), assembled with the text generated from the set of data, machine-encoded in the entry elements below, the Content Creator application MUST handle these two kinds of content, and provide a user interface, which avoids risks of overwriting text automatically derived from the entries with free text typed in by the user (e.g., using forms with dedicated free text areas and distinct protected areas for text generated out of structured data).
- Information that is sent SHALL clearly identify distinctions between:
None
It is known with complete confidence that there are none. This indicates that the sender knows that there is no relevant information of this kind that can be sent.
None Known
None known at this time, but it is not known with complete confidence that none exist.
Asked but unknown
The information was requested but could not be obtained. Used mainly in the context where an observation was made but the result could not be determined.
Unknown
The information is not known, or is otherwise unavailable.
Other, not specified
The actual value does not belong to the assigned value set and is not reported at all by the author.
Other, specify
The actual value does not belong to the assigned value set and the author of the report provides this foreign value anyway.
Not applicable
No proper value is applicable in this context.
Sections that are required to be present but have no information should use one of the above phrases where appropriate in the text element.
Structural elements that are required but have no information shall provide a “nullFlavor” attribute coding the reason why the information is missing.
| Situation | nullFlavor | HL7 Definition |
|---|---|---|
| Asked but unknown | ASKU | Information was sought but not found |
| Unknown | UNK | A proper value is applicable, but not known |
| Other, not specified | OTH | The actual value is not an element in the value domain of a variable. (e.g., concept not provided by required code system). |
| Not applicable | NA | No proper value is applicable in this context |
| Temporarily not available | NAV | Information is not available at this time but it is expected that it will be available later. |
The two situations “None” and “None known” represent effective values, which are part of the related value sets.
The situation “Other, specify” can be handled in two ways in a coded data element:
- Leaving empty the code attribute and providing the non-coded answer in the originalText attribute.
- Providing a value coded from a different coding scheme, when the coding strength of the element is “CWE” (coded with extensions). The attributes code, displayName, codeSystem and codeSystemName then describe the foreign code.
For ancillary techniques, the situation “ not performed” or “none performed” is represented by nullFlavor = NAV.
6.2.1.3 Common structure for CDA APSR
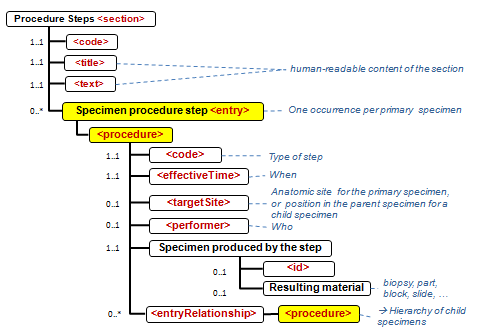
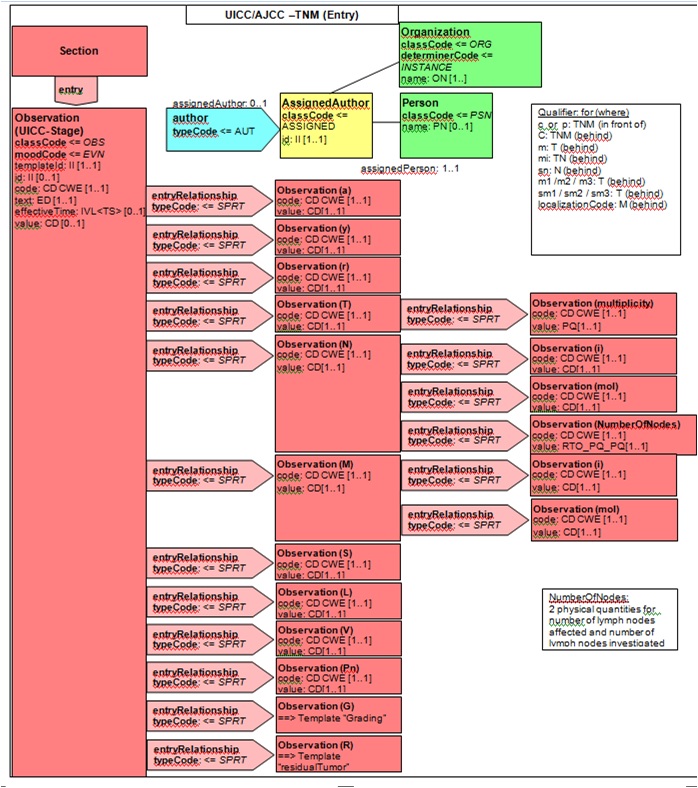
Figure 6.2.1.3-1 summarizes the common structure of the first five CDA APSR Section Content Modules specified here. Regarding the machine-readable part, the figure highlights the organization of entries within a section and of observations within an entry. Specific details such as the structure of sub-sections are not shown on this global picture.
 |
Should these both figures better given in the HL7 Style??? GH |
Figure 6.2.1.3-1 CDA APSR: common structure of machine-readable content for CDA APSR Section Content Modules except Procedure Step Content Module
Figure 6.2.1.3-2 shows the common structure of the Procedure Step Content Module specified here, too.
Figure 6.2.1.3-2 CDA APSR: common structure of machine-readable content for Procedure Step Content Module
Note 1: In order to facilitate a further de-identification process of CDA AP reports for some secondary use (biosurveillance, epidemiology…) the producer of an APSR SHOULD avoid populating any patient identification data (name, sex, birthdate, address …) into the body of the report (neither <entry> elements nor <text> elements). The appropriate location for patient identification data is the CDA header exclusively.
Note 2: The AP sections are those shown on figure 4.1.2.1-1 of Volume 1.
Note 3: The possible sub sections are shown on figure 4.1.2.1-1 of Volume 1.
6.2.2 Common layout for the specification of a CDA Content Module
Each CDA R2 Content Module specified in this Volume is presented with this layout:
6.2.2.1 Content Module Name – OID
Each Content Module is uniquely identified by a unique OID.
6.2.2.1.1 Definition and purpose
This section presents the content module and its purpose.
In case this module is a specialization of a more generic one, the section references its parent template.
6.2.2.1.2 Example
This section delivers a snippet, showing an example of the content module.
6.2.2.1.3 Specification
The form of the specification depends upon the kind of CDA Content Module (document, section, entry, header and/or entry element). It respects the guidelines below:
- The specification provides a table describing the structure of the content module, each element being located through an XPATH expression combined with indentation. The table provides cardinalities, meaning for each elements, references value sets described in section 5 for attributes, and provides the mapping with HL7 V2.5.1 relevant fields. The table also points the content modules nested in the current one, by showing their templateId, and locating their specification in the current PAT TF-3 or in the IHE TF they belong to (PCC, LAB, etc.).
- The table is simplified for section content modules: It only lists the content modules nested in the section template.
- Below the tables appear notes providing additional information and detailing particular constraints on some elements or attributes.
6.2.3 CDA R2 Document Content Modules
6.2.3.1 AP Structured Report (APSR) - 1.3.6.1.4.1.19376.1.8.1.1.1
6.2.3.1.1 Definition and purpose
This Document Content Module defines the base set of constraints that apply to all AP structured report, related to any kind of lesion or diagnostic. In other words, this is the generic template for any AP structured report.
The body of this Document Content Module has the hierarchy of sections and entries depicted by figure 4.1.2.1-1 in Volume 1.
| Id | 2.16.840.1.113883.2.6.60.6.10.1 | Gültigkeit | gültig ab 2014‑05‑13 11:57:57 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Status | Versions-Label | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name | PathologyStructuredReport | Anzeigename | Pathology Structured Report | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Beschreibung | This Document Content Module defines the base set of constraints that apply to all AP structured report, related to any kind of lesion or diagnostic problem. In other words, this is the generic template for any AP structured report. The body of this Document Content Module and of all its specialized children, share a common hierarchy of sections and entries depicted by figure 4.1.2.1-1 in Volume 1 IHE_PAT_Suppl_Rev2 0_TI_2015_05_20 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Klassifikation | CDA Document Level Template | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Offen/Geschlossen | Offen (auch andere als die definierten Elemente sind erlaubt) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Assoziiert mit |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Benutzt von / Benutzt |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Beziehung | Spezialisierung: Template 2.16.840.1.113883.10.12.1 (2005‑09‑07) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Beispiel |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
6.2.4 CDA R2 <section> Content Modules
6.2.4.1 Clinical Information <section> - 1.3.6.1.4.1.19376.1.8.1.2.1
6.2.4.1.1 Definition and Purpose
The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, reason for anatomic pathology procedure, clinical laboratory data, specimen collection procedure including target site, performer, specimen type, specimen(s) clinical description, and tumor site in case of a cancer.
| Hinweis zu dieser Seite |
|---|
Template ClinicalInformationSection
Beschreibung
The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, reason for anatomic pathology procedure, clinical laboratory data, specimen sent for investigation (including collection procedure, target site, performer, specimen type, specimen(s) clinical description, and tumor site in case of a cancer).
Aktuelle Version
Zusammenstellung aller Versionen dieses Templates
6.2.4.2 Intraoperative Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.2
6.2.4.2.1 Definition and Purpose
The Intraoperative Observation section contains an intraoperative diagnosis for each specimen examined, the specimen identification and description, intraoperative observation procedure description (frozen section, gross examination, intraoperative cytology) and derived specimen dissected for other ancillary procedures (flow cytometry, cytogenetics, molecular studies, and electron microscopy).
6.2.4.3 Macroscopic Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.3
6.2.4.3.1 Definition and Purpose
The Macroscopic Observation section contains the description of the specimen(s) received or obtained by the laboratory (specimen type and state), the gross observation, links to gross images, if taken, processing information and tissue disposition (representative sampling and tissue submitted for additional studies or sent to biorepository.
6.2.4.4 Microscopic Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.4
6.2.4.4.1 Definition and Purpose
The Microscopic Observation section contains optionally the histopathologic findings of the case and many laboratories use this section to record the results of histochemical and immunohistochemical stains.
6.2.4.5 Diagnostic Conclusion <section> - 1.3.6.1.4.1.19376.1.8.1.2.5
6.2.4.5.1 Definition and Purpose
The Diagnostic Conclusion section contains diagnoses on all specimens that are delivered to the pathology department from one operation or patient visit to a single clinician on a particular day. The diagnoses for each specimen or group of specimens are reported separately. This section includes additional pathologic finding(s) and the results of ancillary study(ies) and may include diagrams and still images or virtual slides, if taken. In case of cancer, this section includes the cancer checklist.
6.2.4.6 Procedure steps <section> - 1.3.6.1.4.1.19376.1.8.1.2.6
6.2.4.6.1 Definition and Purpose
The Procedure steps section contains the description of tissue dissection: representative specimens and derived specimens dissected for other ancillary procedures (flow cytometry, cytogenetics, molecular studies, electron microscopy, etc.) or biorepository.
6.2.4.7 Report Textual Summary <section> - 1.3.6.1.4.1.19376.1.8.1.2.7
6.2.4.7.1 Definition and Purpose
The Report Textual Summary section is an optional sub-section of the Diagnostic Conclusion section. This section contains a textual summary of the AP report, which can be extracted from the document and inserted into other clinical documents. It addresses the use case where authors of other medical documents feel the need to include a segment such as "...the pathology report states '[...]'", the text content of this section filling the brackets.
6.2.5 CDA R2 <entry> Content Modules
6.2.5.1 Common Specification for all APSR Entry Content Modules
The unique <entry> Content Module available in all the sections except Procedure Steps <section> of the APSR template is Problem (Specimen Information) Organizer.
The unique <entry> Content Module available in Procedure Steps <section> of the APSR template is Specimen Procedure Step
Each <entry> Content Module carries machine-readable data related to one specimen or to one group of specimens observed for this section.
Each <entry> Content Module is repeatable below its section, so as to support the use cases where a section reports on more than one specimen or more than one group of specimens.
6.2.6 CDA R2 Child Element Content Modules
This section specifies the Content Modules designed for child elements. A child element is a child of the CDA header or a child of a <section>, or an element nested at various depths below an <entry>, or an element appearing at some combination of these locations.
6.2.6.1 Specimen Collector in Header – 1.3.6.1.4.1.19376.1.8.1.4.1
6.2.6.1.1 Definition and purpose
This Content Module is usable only in the CDA header.
This Content Module is used only in the situation where the specimen was not collected by the ordering physician. (See use cases in volume 1)
6.2.6.2 Author – 1.3.6.1.4.1.19376.1.8.1.4.2
6.2.6.2.1 Definition and purpose
This Content Module is usable in the CDA header, in a <section> and at various depths of an <entry>.
It describes an author having contributed to the document wholly or to a portion of it (e.g., a section, an observation, a group of observations).
A given document or any delimited portion of it may have more than one author.
An author MAY be a person or a device (manufactured device or software system). In both cases the scoping organization MAY be described.
6.2.6.3 Content Validator – 1.3.6.1.4.1.19376.1.8.1.4.3
6.2.6.3.1 Definition and purpose
This Content Module is usable only in the CDA header.
It describes a pathologist having verified the content of the report, using the element authenticator. This element authenticator is used when the pathologist having verified the content of the report is distinct from the pathologist assuming the legal responsibility for this report, described in the legalAuthenticator element.
The report MAY have zero or more Content Validators.
6.2.6.4 Specimen Information Organizer – 1.3.6.1.4.1.19376.1.8.1.4.4
6.2.6.4.1 Definition and purpose
This Content Module is the unique <entry> available for most of APSR sections (see figure 4.1.2.1-1 in Volume 1). The specimen information organizer organizes information related to the various acts (procedures, observations) performed on a single specimen or group of specimens investigated together.
6.2.6.5 Specimen Collection Procedure generic template – 1.3.6.1.4.1.19376.1.3.1.2
6.2.6.5.1 Definition and purpose
 |
Is this entry really necessary? Its function is better served by the Specimen Procedure Steps entry. GH |
This Content Module is usable within an <entry> element.
This Content Module structures the machine-readable data representing the characteristics of the specimen (identifiers and type) and the procedure that collected it: Type of procedure, time interval, performer (person and organization), approach site, target site.
The “Specimen Collection Procedure” generic template is usable in all <entry> elements of all APSR Document Content Modules.
Each organ-specific APSR Document Content Module mandates an organ-specific template, child of the “Specimen Collection Procedure” generic template. This organ-specific child template has exactly the same structure as the generic one, and brings only a number of vocabulary constraints related to this specific organ:
- The value set bound to the procedure/code element, consisting in a list of triplets (code, codeSystem, displayName) representing the various specimen collection procedures that can be performed on this specific organ.
- The value set bound to the procedure/targetSiteCode element, consisting in a list of triplets (code, codeSystem, displayName) representing the various precise locations for collecting specimens from this specific organ.
Thus, a specimen collection procedure in an <entry> within an organ-specific APSR Document Content Module declares conformance to two templates: The “Specimen Collection Procedure” generic template and the “Specimen Collection Procedure” child template constraining the vocabularies for the contextual organ.
These specimen collection procedure child templates and their attached value sets are provided by the appendix “IHE_PAT_Suppl_APSR_AppendixValue_Sets.xlsx”
6.2.6.6 Informant – 1.3.6.1.4.1.19376.1.8.1.4.6
6.2.6.6.1 Definition and purpose
This Content Module is usable in the CDA header, in a <section> and within an <entry>.
It describes a person having provided some piece of relevant information for the document.
A <ClinicalDocument> or a <section> or any kind of act below an <entry>, MAY have zero or more informant.
6.2.6.7 Additional participant in an entry - 1.3.6.1.4.1.19376.1.8.1.4.7
6.2.6.7.1 Definition and purpose
This Content Module is usable only within an <entry> element.
Additional participants MAY take part in any organizer as well as in any observation of an APSR. These participants MAY be any of these 4:
- Validator: This is the same participation as Content Validator in the header of the report: a pathologist having verified the content (of this particular subset of results).
- Device: A device used to produce this particular subset of results.
- Responsible: The director of a laboratory (described in a performer element at the same level) who produced this particular subset of results.
- Transcriptionist: This is the same participation as dataEnterer in the header of the report: a staff who entered, possibly from dictation, this particular subset of results.
6.2.6.8 Problem Organizer – 1.3.6.1.4.1.19376.1.8.1.4.8
6.2.6.8.1 Definition and purpose
This Content Module is usable within (a Specimen Information Organizer) any Section module.
The problem organizer groups the battery of observations performed to investigate on a single problem identified on a specimen or group of specimens.
6.2.6.9 AP Anatomic pathology Observation template – 1.3.6.1.4.1.19376.1.8.1.4.9
6.2.6.9.1 Definition and purpose
This Content Module is usable within a Problem Organizer.
The “AP Observation”generic template is usable for all AP observations, including ancillary techniques.
Each specific AP observation is associated to a specific template, child of the “AP Observation”generic template. This specific child template has exactly the same structure
as the generic one, and brings only a number of vocabulary constraints related to the type of observation and to the type of organ source of the specimen
observed:
- The code for the specific observation, defined as a value set bound to the observation/code element, containing a single triplet (code,
codeSystem, displayName) representing this specific observation.
- The cardinalities and default type for the observation/value element carrying the results of this observation.
- The domain of values for this observation in case these values are coded. This domain of coded values is defined as a value set bound to the
observation/value element, containing as many triplets (code, codeSystem, displayName) as there are admissible result values for this specific observation performed on a specimen taken from this specific organ.
An AP Observation has a status and an effective time, may describe various participants (persons, devices, organizations), may have a number of additional properties (method, interpretation, text), and may contain embedded images, comments, and sub-observations, which are also AP observations.
6.2.6.10 Embedded Image – 1.3.6.1.4.1.19376.1.8.1.4.10
6.2.6.10.1 Definition and purpose
This Content Module is usable within an <entry> element, in relationship with a display anchor carried in the referencedObject attribute of a <renderMultimedia> element in the <text> element of the <section> holding this <entry>.
The <observationMedia> element carries an image, embedded in B64. This element may be standalone, or encapsulated within a <regionOfInterest> element which defines an overlay
shape to focus on a part of the image.
This <observationMedia> element embeds the image binary data, encoded in B64.
6.2.6.11 Specimen Procedure Step - 1.3.6.1.4.1.19376.1.8.1.4.11
 |
This is an addendum to the reference text of APSR 2.0 GH |
6.2.6.11.1 Definition and purpose
The Specimen Procedure Step <entry> Content Modul contains in machine-readable form all the information concerning one specimen or a group of specimens. It is usable within a Procedure Steps <section> Content Module only.
This Content Module structures the machine-readable data, which characterize the specimens and the procedures of their collection and processing. By nesting via an entryRelationship the entry is used in an iterative manner for each single step of specimen processing, which results in derived (child) specimen in and on the appropriate containers. The resulting specimen is the participant in the procedure according the CDA-RIM (fig. 6.2.6.11.1-1). It carries an ID as well as further attributes, which are defined in the HL7 Specimen DAM HL7 DAM Specimen Release 1.
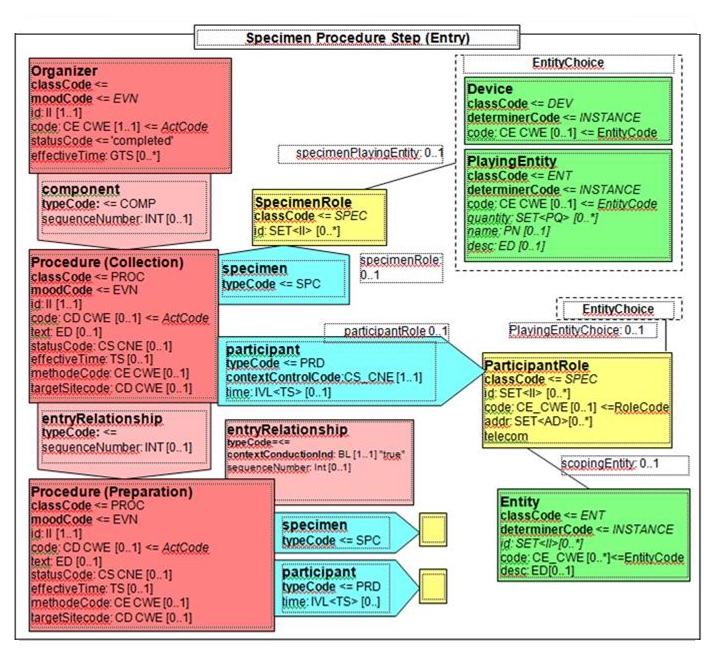
Figure 6.2.6.11.1-1 CDA-RMIM of the Specimen Procedure Steps <entry>
The Code systems and Value sets MAY be organized organ-specific and represented e.g. by PathLex.
6.2.6.12 UICC/AJCC TNM Staging and Grading
 |
This is an addendum to the reference text of APSR 2.0 GH |
6.2.6.12.1 Definition and purpose
The UICC/AJCC TNM <entry> Content Modul contains in machine-readable form all the information concerning a TNM formula for a problem investigated. It is usable within a Problem Organizer <entry> Content Module only.
This Content Module structures the machine-readable data, which characterize the staging and grading of a malignant Tumor according the UICC/AJCC regulations for the TNM system.
Figure 6.2.6.12.1-1 CDA-RMIM of the UICC/AJCC TNM <entry>
6.2.6.13 ICD-O-3 Typing and Grading
 |
This is an addendum to the reference text of APSR 2.0 GH |
6.2.6.13.1 Definition and purpose
The ICD-O-3 Typing and Grading<entry> Content Modul contains in machine-readable form all the information concerning ICD-O-3 Typing and Grading for a problem investigated. It is usable within a Problem Organizer <entry> Content Module only.
This Content Module structures the machine-readable data, which characterize the staging of a tumor or a systemic disease according the WHO regulations for the ICD-O-3 system.
Figure 6.2.6.13.1-1 CDA-RMIM of the ICD-O-3 <entry>
6.2.6.14 Assessment Scales for Scoring systems
 |
This is an addendum to the reference text of APSR 2.0 GH |
6.2.6.14.1 Definition and purpose
The Assessment Scales <entry> Content Modul contains in machine-readable form all the information concerning (semiquantitative) scoring systems often used for describing a tumor grading for a problem investigated. It is usable within the ICD-O-3 Typing and Grading <entry> or within a Problem Organizer <entry> Content Module.
This Content Module structures the machine-readable data, which characterize the grading components of a tumor or a systemic disease according specific regulations.
Figure 6.2.6.14.1-1 CDA-RMIM of an Assessment Scales <entry>