|
|
| (285 dazwischenliegende Versionen von 3 Benutzern werden nicht angezeigt) |
| Zeile 1: |
Zeile 1: |
| | + | __NOTOC__ |
| | <!-- | | <!-- |
| | | | |
| − | Implementierungsleitfaden "Pathologiebefund auf der Basis von CDA R2"
| + | IHE Pathology and Laboratory Medicine Technical Framework Supplement <br/> Anatomic Pathology Structured Report <br/> (APSR) <br/> Rev 2.0 - Draft for Trial Implementation |
| − |
| + | |
| | --> | | --> |
| − | {{Infobox Dokument | + | {{Infobox_Document_IHE |
| − | |Title = Pathologiebefund auf Basis der<br/>HL7 Clinical Document Architecture Release 2<br/>für das deutsche Gesundheitswesen | + | |Dtype = IHE Pathology and Laboratory Medicine<br/>Technical Framework Supplement |
| − | |Short = Pathologiebefund auf der Basis von CDA R2 | + | |Title = Anatomic Pathology Structured Report<br/>(APSR) |
| | + | |Short = IHE PaLM TF Suppl. APSR 2.0 TI |
| | |Namespace = cdapath | | |Namespace = cdapath |
| − | |Type = Implementierungsleitfaden | + | |Type = Rev 2.0 - Draft for Trial Implementation |
| − | |Version = 08 | + | |Version = 091 |
| − | |Date = 13. April 2012 | + | |Submitted = IHE PaLM Technical Committee |
| − | |Copyright = 2010-2012 | + | |Date = 2017-09-27 |
| | + | |Email = palm@ihe.net |
| | + | |Copyright = 2017, IHE International, Inc. |
| | |Status = Draft | | |Status = Draft |
| − | |Period = Abstimmung | + | |Period = Draft |
| − | |OID = n.n. | + | |OID = 1.3.6.1.4.1.19376.1.8.1.1.1 |
| − | |Realm = Deutschland
| + | |Realm = UV |
| − | }}
| |
| − | | |
| − | {{Infobox Ballot Begin}}
| |
| − | {{Ballot | Version = 0.50 | Date = 23.02.2010 | Status = Entwurf | Realm = Deutschland}}
| |
| − | {{Ballot | Version = 0.80 | Date = 15.04.2011 | Status = Entwurf | Realm = Deutschland}}
| |
| − | {{Infobox Ballot End}}
| |
| − | | |
| − | {{Infobox Contributors Begin}}
| |
| − | {{Contributor | Logo = Logo-Agfa.jpg | Name = Agfa HealthCare GmbH | Location = Bonn }}
| |
| − | {{Contributor | Logo = Logo-vivantes.jpg | Name = Vivantes Netzwerk für Gesundheit | Location = Berlin }}
| |
| − | {{Contributor | Name = Projektgruppe „Diagnosen" von HL7 Deutschland | Location = Köln }}
| |
| − | {{Infobox Contributors End}}
| |
| − | | |
| − | | |
| − | HL7 Deutschland e.V.
| |
| − | Geschäftsstelle Köln
| |
| − | An der Schanz 1
| |
| − | 50735 Köln
| |
| − | | |
| − | '''Implementierungsleitfaden'''
| |
| − | | |
| − | '''Pathologie-Befunde auf Basisvon HL7 CDA Rel.2'''
| |
| − | | |
| − | zur Abstimmung durch die Mitglieder von HL7 Deutschland e.V.
| |
| − | | |
| − | Ansprechpartner:
| |
| − | | |
| − | Dr. Frank Oemig, Agfa HealthCare GmbH (Bonn)
| |
| − | | |
| − | '''Dokumentinformation'''
| |
| − | | |
| − | | |
| − | ==Dokumentenhistorie==
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Version!!Stand!!Bearbeiter!!Beschreibung!!Dok.-OID
| |
| − | |-
| |
| − | |06||13.04.12||FO ete.al.||Wikifizierung + Überarbeitung: Neustrukturierung||n.a.
| |
| − | |-
| |
| − | |05||23.02.10||FO||Überarbeitung: Neustrukturierung||n.a.
| |
| − | |-
| |
| − | |04||16.11.09||FO||Überarbeitung: Neustrukturierung||n.a.
| |
| − | |-
| |
| − | |03||15.09.09||FO||Überarbeitung||n.a.
| |
| − | |-
| |
| − | |02||08.07.09||FO||Überarbeitung||n.a.
| |
| − | |-
| |
| − | |01||23.06.09||IR||Dokument erstellt||n.a.
| |
| − | |-
| |
| − | |}
| |
| − | | |
| − | | |
| − | ==Editor==
| |
| − | | |
| − | Dr. Frank Oemig, AGFA HealthCare GmbH, Bonn
| |
| − | | |
| − | | |
| − | ==Autoren==
| |
| − | | |
| − | *Ivonne Riedel, Agfa HealthCare GmbH, Bonn, (FO)
| |
| − | *Dr. Frank Oemig, Agfa HealthCare GmbH, Bonn (IR)
| |
| − | *Prof.Dr. Gunter Haroske, Dresden (GH)
| |
| − | | |
| − | ==Mit Beiträgen von==
| |
| − | | |
| − | Dr. Jochen Thümmler, Vivantes Netzwerk für Gesundheit, Berlin, (JT)
| |
| − | | |
| − | | |
| − | | |
| − | '''Autoren und Copyright-Hinweis, Nutzungs¬hinweise'''
| |
| − | | |
| − | ==Nachnutzungs- bzw. Veröffentlichungsansprüche==
| |
| − | Das vorliegende Dokument wurde von Agfa HealthCaare GbmH, Bonn, und in Kooperation mit der HL7-Benutzergruppe e.V. entwickelt. Die Nachnutzungs- bzw. Veröffentlichungs¬ansprüche sind nicht beschränkt.
| |
| − | | |
| − | | |
| − | Der Inhalt dieser Spezifikation ist öffentlich.
| |
| − | | |
| − | Zu beachten ist, dass Teile dieses Dokuments auf dem Abstimmungspaket 2 vom 17.Mai 2009 und der Normative Edition 2008 von HL7-Version 3 beruhen, für die © Health Level Seven, Inc. gilt.
| |
| − | Näheres unter [http://www.h7.de-http://www.h7.de/] und [http://www.hl7.org-http://www.hl7.org/].
| |
| − | | |
| − | Die Erweiterung oder Ablehnung der Spezifikation, ganz oder in Teilen, ist dem Vorstand der Benutzergruppe und den Editoren/Autoren schriftlich anzuzeigen.
| |
| − | | |
| − | | |
| − | Alle auf nationale Verhältnisse angepassten und veröffentlichten HL7-Spezifkationen können ohne Lizenz- und Nutzungsgebühren in jeder Art von Anwendungssoftware verwendet werden.
| |
| − | | |
| − | | |
| − | | |
| − | | |
| − | Disclaimer
| |
| − | | |
| − | Obwohl diese Publikation mit größter Sorgfalt erstellt wurde, kann weder HL7 Deutschland e.V. noch die an der Erstellung beteiligten Firmen keinerlei Haftung für direkten oder indirekten Schaden übernehmen, die durch den Inhalt dieser Spezifikation entstehen könnten.
| |
| − | | |
| − | | |
| − | | |
| − |
| |
| − | | |
| − | Inhaltsverzeichnis Seite
| |
| − | | |
| − | | |
| − | | |
| − | =Einleitung=
| |
| − | | |
| − | | |
| − | | |
| − | | |
| − | | |
| − | ==Einleitung==
| |
| − | Dieses Dokument enthält einen ersten Entwurf für die Umsetzung von Pathologie-Berichten mit Hilfe von HL7 CDA R2. Exemplarisch soll diese Entwicklung für die Pathologieintegration innerhalb des Vivantes Netzwerks für Gesundheit, Berlin, prototypisch genutzt werden.
| |
| − | | |
| − | Orientiert wird dabei auf eine möglichst vollständige Berücksichtigung des "Leitfadens Pathologie/Neuropathologie (ehem. TM-30)" des Sektorkomitees Pathologie für die Anwendung der DIN EN ISO/IEC 17020 in der Pathologie/Neuropathologie.
| |
| − | | |
| − | Weiterhin wird angestrebt, die durch den Bundesverband Deutscher Pathologen und die Deutsche Gesellschaft für Pathologie veröffentlichten "Empfehlungen zur pathologisch-anatomischen Diagnostik von Kolorektalen Karzinomen, Mammakarzinomen und Prostatakarzinomen" in HL7 CDA R2 kompatible Templates zur Integration in Pathologie-Management-Systeme umzusetzen.
| |
| − | | |
| − | Auf dieser Basis der soll der Import von HL7 CDA R2 Dokumenten von der Pathologie in ORBIS umgesetzt werden.
| |
| − | | |
| − | ==3. Grundlage==
| |
| − | Grundlage dieses Konzeptes ist der Implementierungsleitfaden der VHitG für den Arztbrief des deutschen Gesundheitswesens sowie der Diagnose- und Datentypleitfaden.
| |
| − | | |
| − | * VHitG Arztbrief, v1.5, \[CDAr2Arztbrief\]
| |
| − | * Diagnoseleitfaden v0.99b, 13.12.09
| |
| − | * Datentypleitfaden
| |
| − | | |
| − | {{NoteBox|
| |
| − | Anm.: Bei IHE Anatomic Pathology ist derzeit ein neues Profil in Arbeit: "Anatomic Pathology reporting to Public Health" ([www.ihe.net-http://www.ihe.net/]). Hierbei handelt es sich jedoch um HL7 v2.5 ORU Nachrichten, so dass die Inhalte nicht weiter genutzt werden können.
| |
| − | }}
| |
| − | | |
| − | ==4. Disclaimer==
| |
| − | Dieses Dokument enthält keine komplette Spezifikation eines HL7 CDA R2 Arztbriefes bzw. Dokumentes. Es werden Teile eines Arztbriefes spezifiziert, wie er im Rahmen der Pathologieintegration innerhalb der Vivantes Gruppe benötigt werden. Ziel dieser Integration soll es sein, alle für die onkologische Tumordokumentation relevanten Daten in ORBIS zu importieren. Eine Vollständigkeit des Arztbriefes kann daher nicht gewährleistet werden.
| |
| − | | |
| − | Weiterhin wird nur eine unidirektionale Kommunikation des HL7 CDA Arztbriefes spezifiziert – Import nach ORBIS.
| |
| − | | |
| − | | |
| − | =Dynamisches Modell=
| |
| − | | |
| − | ==Übersicht==
| |
| − | Hier muss das dynamische Modell hin. Derzeit sieht das relativ einfach aus:
| |
| − | | |
| − | [[file:Cdapat_dynamisches_modell.gif|500px|dynamisches Modell]]
| |
| − | | |
| − | Abbildung 1: dynamisches Modell
| |
| − | | |
| − | =Statisches Modell=
| |
| − | | |
| − | | |
| − | ==Übersicht==
| |
| − | In diesem Abschnitt wird grob der Aufbau und die Struktur von HL7 CDA R2 Dokumenten erläutert (entnommen aus dem Implementierungsleitfaden Kapitel 3).
| |
| − | | |
| − | Wie alle Spezifikationen von Nachrichten in HL7 basiert auch die Clinical Document Architecture auf dem RIM und ist als HL7 V3 Modell repräsentiert. Grob gesprochen besteht ein CDA Dokument aus einem '''Header''' und einem '''Body''', der wiederum '''Body Structures''' und '''Body Entries''' aufweist. An die Entries können externe Referenzen ('''External References''') geknüpft sein. Der folgende Überblick zeigt die Hauptkomponenten des CDA R2 Modells auf und in der folgenden Abbildung ist das Ganze in XML-artiger Darstellung gezeigt.
| |
| − | | |
| − |
| |
| − | [[file:Cdapat_cda_rmim.jpg|600px|CDA RMIM]]
| |
| − | | |
| − | Abbildung 2: CDA-RMIM (vereinfachte Darstellung)
| |
| − | | |
| − | Die nachfolgende vereinfachte Graphik zeigt die Darstellung in XML:
| |
| − | | |
| − | [[file:Cdaab1_xml_gesamt.png|600px|CDA Level 3 Entries]]
| |
| − |
| |
| − | Abbildung 3: CDA Level 3 Entries (vereinfachter Ausschnitt)
| |
| − | | |
| − | Die Informationen zum Patienten, zum Dokument selbst, zu den weiteren beteiligten Personen und Organisationen sowie der dokumentierten Episode (Zeitereignisse) sind zum '''CDA Header''' zusammengefasst, hochstrukturiert und von der Semantik her festgelegt.
| |
| − | | |
| − | Die Informationen im Header unterstützen einen Austausch klinischer Dokumente über Institutionsgrenzen hinweg. Er trägt Informationen über das Dokument selbst (eine eineindeutige Identifikation, eine Andeutung des Typs des Dokuments), über „Teilnehmer" am Dokument (an der Dokumentation beteiligte Heilberufler, Autoren, und natürlich den Patienten selbst), sowie über Beziehungen zu Dokumenten (zu Anforderungen und anderen Dokumenten). Mit den Informationen des Headers werden Dokumentenmanagementsysteme unterstützt, der Header stellt dafür entsprechende Mechanismen zur Verfügung. Schließlich hat man mit den im CDA Header verfügbaren Informationen die Zusammenführung einer individuellen (lebenslangen) Patientenakte vor Augen.
| |
| − | | |
| − | ==Gesamtstruktur==
| |
| − | | |
| − | [[file:Cdapat_cda_gesamt.gif|400px|CDA Gesamtstruktur]]
| |
| − | | |
| − | Abbildung 4: Gesamtstruktur
| |
| − | | |
| − | | |
| − | ===Dokumenttypen===
| |
| − | ''Diese Tabelle sollte noch einmal diskutiert werden, da die Struktur nicht logisch erscheint. Sektionsbefunde werden i.A. stark abweichend von histologischen Befunden aufgebaut und sollten deshalb nicht prioritär behandelt werden. Eine Abgrenzung von Ektomien ist nicht sehr sinnvoll, da es nur eine Materialkategorie darstellt. Diese können von Fragestellung zu Fragestellung sehr variieren. Also sollte man den Einstieg allgemein halten und erst nach dem diagnostischen problem diversifizieren in z.B. Biopsie (wiederum in verschiedenen technischen Varianten, z.B. Stanzbiopsie mit unterschiedlicher Guidance, Nadelstärke und Technik), Resektion (Ektomien, wie Prostatektomien, Mastektomien etc., Resektionen wie Segmentresektion, Exzision wie TME (Totale mesorektale Exzision), um nur einige typische aus den angepeilten Tumorpathologien zu nennen. Das lässt sich aber organspezifisch alles im Materialteil darstellen und auch kodieren)
| |
| − | Die Grundstruktur eines Befundes ist: Material-Makroskopische Beurteilung-Mikroskopische Beurteilung-Diagnose/Zusammenfassung.
| |
| − | Die Klinische Fragestellung wird von einigen Kollegen in den Befund übernommen, sinnvoll wäre ihre Berücksichtigung bei einer bidirektionalen Verbindung zum KIS.
| |
| − | Immunhistologischer und molekularpathologischer Befund sind Ergebnis der pathologischen Stufendiagnostik und in der Regel im Befund eingearbeitet. Man könnte über eine separate, sehr methodisch orientierte Extension als Befundanhang diskutieren.''
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Dokumenttyp <br>Abschnitt!!Obduktion/ Sektion!!Ektomie!!histologische Untersuchung!!molekularbiologische Untersuchung
| |
| − | |-
| |
| − | |Anrede|| <nowiki>[0..1 ]</nowiki>||<nowiki>[0..1]</nowiki>||<nowiki>[0..1]</nowiki>||<nowiki>[0..1]</nowiki>
| |
| − | |-
| |
| − | |Vorbefunde||<nowiki>[0..1]</nowiki>||\[0..1\]||||
| |
| − | |-
| |
| − | |Indikation||||||||
| |
| − | |-
| |
| − | |Fragestellung||\[0..1\]||\[0..1\]||||
| |
| − | |-
| |
| − | |Todesursache||\[1..1\]||\[0..1\]||||
| |
| − | |-
| |
| − | |Vorgeschichte||||||||
| |
| − | |-
| |
| − | |Grund¬erkrankungen||\[1..1\]||||||
| |
| − | |-
| |
| − | |Material¬aufbereitung||||||||
| |
| − | |-
| |
| − | |Makros¬kopie||\[0..1\]||||\[0..1\]||\[0..1\]
| |
| − | |-
| |
| − | |Intraoperativer Schnellschnitt||||||\[0..1\]||
| |
| − | |-
| |
| − | |Präparat¬radiographie||||||||
| |
| − | |-
| |
| − | |Mikros¬kopie||\[0..1\]||||\[0..1\]||\[0..1\]
| |
| − | |-
| |
| − | |Immunhistologie||\[0..1\]||||||
| |
| − | |-
| |
| − | |Elektronen¬mikroskopie||\[0..1\]||||||
| |
| − | |-
| |
| − | |molekularpatholog. Befund||\[0..1\]||||||\[1..1\]
| |
| − | |-
| |
| − | |Epikrise||\[0..1\]||||||
| |
| − | |-
| |
| − | |ausführl. kritische gutachterliche Stellungnahme/ Befund||\[1..1\]||||\[1..1\]||\[1..1\]
| |
| − | |-
| |
| − | |Färbungen||\[0..1\]||||\[0..1\]||\[0..1\]
| |
| − | |-
| |
| − | |Kommentar||||||||
| |
| − | |-
| |
| − | |Verschlüsselung/ Stadium/spezielle Schlüssel||||||||
| |
| − | |-
| |
| − | |zusammenfassende Beurteilung||||||||
| |
| − | |-
| |
| − | |Weitergabemodus||\[0..1\]||\[0..1\]||\[0..1\]||\[0..1\]
| |
| − | |-
| |
| − | |Gruß||\[0..1\]||||\[0..1\]||\[0..1\]
| |
| − | |-
| |
| − | |Anlagen||||||||
| |
| − | |-
| |
| − | |immunhistolog. Tabelle||||||||
| |
| − | |-
| |
| − | |molekularpatholo¬gische Tabelle||||||||
| |
| − | |-
| |
| − | |Graduierungs¬schemata||||||||
| |
| − | |-
| |
| − | |Attribut-Wert-Paare||\[0..1\]||||\[0..1\]||\[0..1\]
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 1: Dokumenttypen und deren Inhalt
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Abschnitt!!LOINC!!Beschreibung!!Level
| |
| − | |-
| |
| − | |Anrede||||||2
| |
| − | |-
| |
| − | |Vorbefunde||||||
| |
| − | |-
| |
| − | |Fragestellung||||||2
| |
| − | |-
| |
| − | |Todesursache||||||
| |
| − | |-
| |
| − | |Vorgeschichte||||||
| |
| − | |-
| |
| − | |Grunderkrankungen||||||
| |
| − | |-
| |
| − | |Materialaufbereitung||||||
| |
| − | |-
| |
| − | |Makroskopie||22634-0||||
| |
| − | |-
| |
| − | |Intraoperativer Schnellschnitt|||||
| |
| − | |-
| |
| − | |Präparatradiographie||||||
| |
| − | |-
| |
| − | |Mikroskopie||22635-7||||
| |
| − | |-
| |
| − | |Immunhistologie||||||
| |
| − | |-
| |
| − | |Elektronenmikroskopie||||||
| |
| − | |-
| |
| − | |molekularpatholog. Befund||||||
| |
| − | |-
| |
| − | |Epikrise||33746-9||||
| |
| − | |-
| |
| − | |kritische gutachterliche Stellungnahme||||||
| |
| − | |-
| |
| − | |Diagnosen||22637-3||||3
| |
| − | |-
| |
| − | |Beurteilung||22034-3||||
| |
| − | |-
| |
| − | |Kommentar||||||2
| |
| − | |-
| |
| − | |Verschlüsselung/Stadium/ spezielle Schlüssel||||||
| |
| − | |-
| |
| − | |Weitergabemodus||||||2
| |
| − | |-
| |
| − | |Gruß||||||2
| |
| − | |-
| |
| − | |Anlagen||||||2
| |
| − | |-
| |
| − | |immunhistolog. Tabelle||||||2
| |
| − | |-
| |
| − | |molekularpath. Tabelle||||||2
| |
| − | |-
| |
| − | |Graduierungsschemata||||||2
| |
| − | |-
| |
| − | |Attribut-Wert-Paare||||||3
| |
| − | |-
| |
| − | |Färbungen||||||3
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 2: LOINC-Codes für die Sektionen
| |
| − | | |
| − | ==CDA-Header==
| |
| − | Alle XML Arztbriefe beginnen mit dem Wurzelelement ''ClinicalDocument'' und der vorgeschriebene Zeichensatz ist UTF-8.
| |
| − | | |
| − | Daraus ergibt sich folgende Struktur, die wie aufgeführt umzusetzen ist. Dabei sind '''fett''' gedruckte Bereiche unverändert einzubauen.
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <?xml version="1.0"? encoding="UTF-8">
| |
| − | <ClinicalDocument
| |
| − | xmlns="urn:hl7-org:v3"
| |
| − | xmlns:voc="urn:hl7-org:v3/voc"
| |
| − | xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance">
| |
| − | <typeId root="2.16.840.1.113883.1.3" extension="POCD_HD000040"/>
| |
| − | | |
| − | <!-- CDA Header -->
| |
| − | ... siehe Beschreibung CDA R2 Header
| |
| − | | |
| − | <!-- CDA Body -->
| |
| − | <component>
| |
| − | <structuredBody>
| |
| − | ... siehe Beschreibung CDA R2 Body
| |
| − | </structuredBody>
| |
| − | </component>
| |
| − | </ClinicalDocument>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | In diesem Abschnitt werden die Elemente des CDA Headers erläutert, die zwingend in den CDA HL7 R2 Arztbrief einzubinden sind.
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = elm
| |
| − | | rim = act
| |
| − | | name = document
| |
| − | | desc = Document
| |
| − | | dt =
| |
| − | | card = 1..1
| |
| − | | conf = M
| |
| − | }}
| |
| − | | |
| − | ===Dokumenten-ID===
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = att
| |
| − | | rim = act
| |
| − | | name = id
| |
| − | | desc = Dokumenten-ID
| |
| − | | dt = II
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | Jeder Arztbrief muss genau eine eindeutige DokumentenID aufweisen. Diese DokumentenID identifiziert ein Dokument weltweit und für alle eindeutig.
| |
| − | Diese muss folgendermaßen aussehen:
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <id extension="13234453645" root="2.16.840.1.113883.2.6.15.3.427.1"/>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | Das ''@extension'' Attribut enthält eine eindeutige Dokumentennummer, die von der in ''@root'' genannten Authority vergeben wird. Im ''@root'' Attribut wird das Dokument-erzeugende Anwendungssystem über eine OID<sup> </sup> identifiziert:
| |
| − | Für die Kommunikation nach außen muss eine OID gewählt werden, die eindeutig für die Instanz des Anwendersystems ist. In der Regel werden diese OIDs vom Hersteller des jeweiligen Anwendersystems kommen, der seine tatsächlichen Installationen (Applikations-Instanzen) mit entsprechenden eindeutigen OIDs zu versehen hat. Das heißt, dass jede Installation eines Anbieters eine eindeutige OID besitzt und verwendet.
| |
| − | | |
| − | ===Typisierung des Dokuments===
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = att
| |
| − | | rim = act
| |
| − | | name = code
| |
| − | | desc = Typ des Dokuments
| |
| − | | dt = CE CWE
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | Über das @code Attribut wird eine Typisierung des Dokuments vorgenommen.
| |
| − | | |
| − | Im Falle der Integration des Pathologiesystems von Vivantes ist folgender Eintrag zu verwenden.
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Dokumenten-Typ!!Deutsche Bezeichnung!!Berufsgruppe!!Umgebung
| |
| − | |-
| |
| − | |11529-5||Surgical pathology report||Pathologischer Bericht|| ||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 3: LOINC-Codes für Dokumenttypen (OID 2.16.840.1.113883.6.1)
| |
| − | | |
| − | Weitere Typen sind bei Interesse dem Implementierungsleitfaden der VHitG S. 46 Tabelle 3 zu entnehmen.
| |
| − | | |
| − | Für das ''@code'' Attribut wird das LOINC Codesystem verwendet. Es muss das ''@codeSystem'' Attribut daher mit dem OID des LOINC gefüllt werden.
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <code code="11529-5" codeSystem="2.16.840.1.113883.6.1"/>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ===Titel===
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = att
| |
| − | | rim = act
| |
| − | | name = title
| |
| − | | desc = Title des Dokuments
| |
| − | | dt = ST
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <title>Pathologisch anatomische Begutachtung [mit kritischer Stellungnahme]</title>
| |
| − | </syntaxhighlight>
| |
| − | | |
| − | ===Erstellungsdatum===
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = att
| |
| − | | rim = act
| |
| − | | name = effectiveTime
| |
| − | | desc = Erstellungsdatum
| |
| − | | dt = TS
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | Das ''@effectiveTime'' Attribut enthält das Erstellungdatum des Dokumentes. Es muss mindestens eine Jahres-, Montats- und Tagesangabe enthalten. Eine Stunden- und Minutenangabe ist optional.
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <effectiveTime value="200509241634"/>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ===Teilnehmende Parteien===
| |
| − | Innerhalb eines CDA Dokumentes müssen verschiedene teilnehmende Parteien aufgeführt werden.
| |
| − | | |
| − | ====Patient====
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = elm
| |
| − | | rim = part
| |
| − | | name = recordTarget
| |
| − | | desc = Patient
| |
| − | | dt =
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | In diesem Abschnitt im CDA Header wird der Patient beschrieben/erfasst. Dieser setzt sich zusammen aus einer Patientenrolle sowie dem Patienten'' ''selbst. Diese werden im ''recordTarget'' zusammengeführt.
| |
| − | | |
| − | <u>Patientenrolle</u>
| |
| − | Im CDA-Header muss mindestens eine Patientenrolle beschrieben sein, die genau von einer Person gespielt wird.
| |
| − | | |
| − | Verpflichtend muss in diesem Bereich die Patientenidentifikationsnummer angegeben werden. Diese setzt sich zusammen aus dem @extension Attribut, das die ID des Patienten enthält sowie dem @root Attribut, das die OID des Systems enthält, das die ID vergeben hat.
| |
| − | | |
| − | Ein Beispiel muss folgendermaßen aussehen:
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <id extension="6245" root="2.16.840.1.113883.3.933"/>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | <u>Patient</u>
| |
| − | Die Rolle des Patienten wird durch eine Person gespielt.
| |
| − | | |
| − | In dem Attribut ''@name<sup> </sup>'' ist der Name des Patienten untergebracht. Der Name wird wiederrum unterteilt in die ''@given'' und ''@family'' Attribute, die den Vornamen und den Familiennamen des Patienten enthalten.
| |
| − | | |
| − | Ein kompletter ''recordTarget'' ist im Folgenden angegeben.
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <recordTarget>
| |
| − | <!--- Patienten-Daten -->
| |
| − | <patientRole>
| |
| − | <id extension="6245" root="2.16.840.1.113883.3.933"/>
| |
| − | <patient>
| |
| − | <name>
| |
| − | <prefix>Dr.</prefix>
| |
| − | <given>Paul</given>
| |
| − | <family>Pappel</family>
| |
| − | </name>
| |
| − | </patient>
| |
| − | </patientRole>
| |
| − | </recordTarget>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ====Autor====
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = elm
| |
| − | | rim = part
| |
| − | | name = author
| |
| − | | desc = Autor
| |
| − | | dt =
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | Neben dem Patienten muss ein Autor (''author'') angegeben werden, welcher das Dokument verfasst hat.
| |
| − | | |
| − | <u>Time</u>
| |
| − | Im verpflichtend anzugebenden ''@time'' Attribut wird der Zeitpunkt der Dokumentation angegeben.
| |
| − | | |
| − | <u>Autor</u>
| |
| − | Informationen über den Autor werden in der ''assignedAuthor'' Klasse angegeben.
| |
| − | | |
| − | <id extension="190388km89" root="2.16.840.1.113883.3.24535"/>
| |
| − | | |
| − | <u>Name</u>
| |
| − | In dem Attribut ''@name'' ist der Name des Autors untergebracht. Der Name wird wiederrum unterteilt in die ''@given'' und ''@fami''ly Attribute, die den Vornamen und den Familiennamen des Autors enthalten.
| |
| − | | |
| − | Ein kompletter ''author'' ist im Folgenden angegeben.
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <author>
| |
| − | <time value="20050829"/>
| |
| − | <assignedAuthor>
| |
| − | <id extension="190388km89" root="2.16.840.1.113883.3.24535"/>
| |
| − | <assignedPerson>
| |
| − | <name>
| |
| − | <prefix>Dr.med.</prefix>
| |
| − | <given>Theo</given>
| |
| − | <family>Phyllin</family>
| |
| − | </name>
| |
| − | </assignedPerson>
| |
| − | </assignedAuthor>
| |
| − | </author>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ====Verwaltende Organisation====
| |
| − | | |
| − | {{AttDesc
| |
| − | | ae = elm
| |
| − | | rim = part
| |
| − | | name = custodian
| |
| − | | desc = erwaltende Organisation
| |
| − | | dt =
| |
| − | | card = 1..1
| |
| − | | conf =
| |
| − | }}
| |
| − | | |
| − | | |
| − | Die Organisation (''custodian''), die für die Verwaltung des Dokuments verantwortlich ist, muss verpflichtend in der entsprechenden Klasse wiedergegeben werden.
| |
| − | Die Organisation muss mindestens mit einer ID gekennzeichnet werden.
| |
| − | | |
| − | Ein kompletter ''custodian'' ist im Folgenden angegeben.
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <custodian>
| |
| − | <assignedCustodian>
| |
| − | <representedCustodianOrganization>
| |
| − | <id extension="175648374" root="1.2.276.0.76.4.5">
| |
| − | <name>
| |
| − | ...
| |
| − | </name>
| |
| − | </representedCustodianOrganization>
| |
| − | </assignedCustodian>
| |
| − | </custodian>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ==17. CDA Body==
| |
| − | Die eigentliche klinische Dokumentation wird im so genannten '''CDA Body''' festgehalten. Im Vordergrund steht hier „lesbarer" (narrativer) Text, der verpflichtender Bestandteil von CDA R2 Dokumenten ist und die Interoperabilität zwischen den menschlichen Kommunikationspartnern garantiert.
| |
| − | Hier sind Möglichkeiten gegeben, diesen Text grob zu strukturieren, wie man dies von den Möglichkeiten der Textverarbeitung her kennt. Zur Strukturierung stellt die Standardspezifikation eine Reihe von XMLElementen zur Verfügung, die als Body Structures zusammengefasst werden können. Der Body enthält ein oder mehrere Abschnitte (sections). Diese können auch ineinander geschachtelt sein, so wie Kapitel und Unterkapitel in einem Buch. Zudem sind Strukturierungen im Sinne von Tabellen oder Listen möglich.
| |
| − | | |
| − | * Abschnitte <section>
| |
| − | * Paragrafen <paragraph>
| |
| − | * Kennzeichnung von bestimmten Inhalten <content>
| |
| − | * Überschriften <caption>
| |
| − | * Tabellen <table>
| |
| − | * Listen <list>
| |
| − | | |
| − | Sections enthalten immer einen narrativen Block und erfüllen damit eine der oben genannten Maximen von CDA: die Mensch-zu-Mensch-Interoperabilität, die Lesbarkeit der Informationen für den Menschen. Im narrativen Block, durch das Textattribut in der section-Klasse repräsentiert, wird eingebetteter Text innerhalb eines Abschnittes angegeben. Dabei kann mit oben genanntem <content> Element bestimmter Inhalt gesondert gekennzeichnet werden.
| |
| − | | |
| − | Zusammengefasst werden im Textblock (teils so auch schon in CDA Release 1 realisiert) u.a. folgende Möglichkeiten der Struktur- und Formgebung des fließenden Textes gegeben:
| |
| − | | |
| − | * Zeilenumbrüche <br>
| |
| − | * Stilistische Angaben (unterstreichen, fett, kursiv etc.)
| |
| − | * Hoch- und Tiefstellung von Text
| |
| − | * Fußnoten
| |
| − | * Symbole
| |
| − | * Revisionsmarken im Text wie <delete>, <insert>
| |
| − | | |
| − | Mit den beschriebenen Body Strukturen können '''CDA Entries''' verbunden sein. Diese repräsentieren den „computerlesbaren Teil" innerhalb eines Dokumentenabschnitts. Body Entries sind im Prinzip eine Auswahl aus Klassen mitsamt Attributen aus dem HL7 Referenz-Informationsmodell (RIM).
| |
| − | | |
| − | ===Modell===
| |
| − | Nachfolgend ist das CDA-Modell angegeben, so wie es für den Pathologie-Bericht instanziiert wird:
| |
| − |
| |
| − | [[file:Cdapat_level3_modell.gif|400px|Level 3Modell]]
| |
| − | | |
| − | Abbildung 5: Level-3-Modell
| |
| − | | |
| − | | |
| − | ===Abschnitte ("Sections")===
| |
| − | Im folgenden sollen die einzelnen Abschnitte näher spezifiziert werden.
| |
| − | | |
| − | ====Anrede====
| |
| − | Dies ist bereits Bestandteil des VHitG-Arztbriefes.
| |
| − | | |
| − | ====Vorbefunde====
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ====Fragestellung====
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ====Todesursache====
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ====Vorgeschichte====
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ====Grunderkrankungen====
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ====Materialaufbereitung====
| |
| − | | |
| − | Für diese Section ist durch IHE APSR vorgeschlagen, "Procedure Steps" zu verwenden (OID 1.3.6.1.4.1.19376.1.8.1.2.6), die die verwendeten (Routine- und) Spezialmethoden an der repräsentativen gewebsprobe oder ihren repräsentativen Teilen beschreiben. Als ein Template soll es in einem entry-Element eingesetzt werden. Für den Prozedurencode und den target site code gibt es vorgeschlagene value sets.
| |
| − | | |
| − | Dieser Vorschlag sollte in deutscher Übersetzung übernommen werden.
| |
| − | | |
| − | ====Makroskopie====
| |
| − | | |
| − | | |
| − | ====Intraoperativer Schnellschnitt====
| |
| − | | |
| − | | |
| − | ''Für diese Section ist durch IHE APSR vorgeschlagen, "Intraoperative Observation" zu verwenden (OID 1.3.6.1.4.1.19376.1.8.1.2.2), die die intraoperative Diagnose für jede beurteilte Probe einschl. Proben-und Prozedurbeschreibung beschreibt. Ein maschinenlesbares Entry-Modul "Specimen Intraoperative Observation Entry", Template ID 1.3.6.1.4.1.19376.1.8.1.3.2 sowie ein Modul "Author of the section", Template ID 1.3.6.1.4.1.19376.1.8.1.4.2 innerhalb dieser Section ist vorgesehen. Für die Kodierung von 1.3.6.1.4.1.19376.1.8.1.3.2 ist LOINC vorgesehen, ein Ergebnis einer Anfrage am Regenstrief-Institut steht aber noch aus.''
| |
| − | | |
| − | ''Dieser Vorschlag sollte in deutscher Übersetzung angepasst werden.''
| |
| − | | |
| − | ''Außerdem sollte ein weiteres Entry-Modul für Prozessdaten generiert werden, das Probeneingang (Specimen->SpecimenProcessStep), Ende der Probenuntersuchung (documentationOfServiceEvent->Specimen), Ende der Befundung (Specimen->ObservationEventauthor.time) oder Übermittlung Befund erfasst und zusätzlich die Diagnosequalität nach Abschluss der Untersuchungen in eine der vier Kategorien einteilt: Richtig Positiv, Richtig Negativ, Falsch Positiv, Falsch Negativ hinsichtlich der fast ausschließlich vorliegenden Fragestellung "Malignität?" Für diese Kategorien gibt es offensichtlich noch keine Kodierungen (außer UMLS und SNOMED CT, hier "Modifier mainly for procedure (qualifier value), Concept ID 106239005)??''
| |
| − | | |
| − | ====Präparatradiographie====
| |
| − | | |
| − | sollte als eine mögliche Procedure step geführt werden
| |
| − | | |
| − | ====Mikroskopie====
| |
| − | | |
| − | | |
| − | ====Immunhistologie====
| |
| − | | |
| − | | |
| − | ====Elektronenmikroskopie====
| |
| − | | |
| − | | |
| − | ====Molekularpathologischer Befund====
| |
| − | | |
| − | | |
| − | ====Epikrise====
| |
| − | Dies ist bereits Bestandteil des VHitG-Arztbriefes.
| |
| − | | |
| − | ====Diagnosen====
| |
| − | Die Diagnosen sind gemäß Diagnoseleitfaden zu übermitteln!
| |
| − | Die Darstellung wird aus den codierten Informationen abgeleitet.
| |
| − | | |
| − | Trotzdem sollte hier noch ein vollständiges Beispiel angeführt werden!
| |
| − | | |
| − | | |
| − | ====Färbungen====
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Bedeutung!!Datentyp!!OID
| |
| − | |-
| |
| − | |Antikörper (Kurzbezeichnung)||String||??
| |
| − | |-
| |
| − | |Färbung||Code||??
| |
| − | |-
| |
| − | |Reaktion||Code||??
| |
| − | |-
| |
| − | |Prozent||Coded Ordinal||??
| |
| − | |-
| |
| − | |Verteilung||Code||??
| |
| − | |-
| |
| − | |Fixierung||Code||??
| |
| − | |-
| |
| − | |Gewebe||Code||??
| |
| − | |}
| |
| − | Tabelle 4: Färbungen
| |
| − | | |
| − | =====Text-Beispiel=====
| |
| − | Nachfolgend ein Beispiel in der Text-Darstellung:
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Antikörper!!Färbung!!Reaktion!!Proz.!!Verteilung!!Fixierung!!Gewebe
| |
| − | |-
| |
| − | |IF Ep MNF116||positiv||stark||||diffus||Formalin||Tumor isolierte Tumor¬zelle
| |
| − | |-
| |
| − | |Kontrolle Negativ||negativ||keine||||||Formalin||Kontroll¬gewebe
| |
| − | |-
| |
| − | |IF Ep CK 05/6||negativ||keine||||||Formalin||DCIS
| |
| − | |-
| |
| − | |Rez Oestrogen||positiv||mittel||60||diffus||Formalin||Kontroll¬gewebe
| |
| − | |-
| |
| − | |Rez Oestrogen||positiv||mittel||||diffus||Formalin||Tumor
| |
| − | |-
| |
| − | |Rez Progesteron||negativ||keine||||||Formalin||Kontroll¬gewebe
| |
| − | |-
| |
| − | |Rez Progesteron||positiv||stark||||||Formalin||Tumor
| |
| − | |-
| |
| − | |Prol Ki-67||nicht auswertbar||||||||Formalin||
| |
| − | |-
| |
| − | |TM E-Cadherin||positiv||stark||||||Formalin||Tumor
| |
| − | |-
| |
| − | |TM Oncoprotein C-erbB-2||negativ||keine||||diffus||Formalin||Tumor
| |
| − | |-
| |
| − | |IF Ep CK 18||positiv||mittel||||diffus||Formalin||DCIS
| |
| − | |-
| |
| − | |TM Oncoprotein C-erbB-2||positiv||stark||||diffus||Formalin||Kontroll¬gewebe
| |
| − | |-
| |
| − | |IF Ep MNF116||negativ||keine||||||Formalin||Lymph¬knoten
| |
| − | |-
| |
| − | |}
| |
| − | | |
| − | | |
| − | =====Abbildung in CDA=====
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <section>
| |
| − | <!-- Darstellung als Tabelle -->
| |
| − | <text>
| |
| − | <tbody>
| |
| − | <tr>
| |
| − | <th>Antikörper</th>
| |
| − | <th>Färbung</th>
| |
| − | <th>Reaktion</th>
| |
| − | <th>Prozent</th>
| |
| − | <th>Verteilung</th>
| |
| − | <th>Fixierung</th>
| |
| − | <th>Gewebe</th>
| |
| − | </tr>
| |
| − | <tr>
| |
| − | <td><content ID="d1">IF Ep MNF116</content></td>
| |
| − | <td><content ID="d2">positiv</content></td>
| |
| − | <td><content ID="d3">stark</content></td>
| |
| − | <td><content ID="d4"> </content></td>
| |
| − | <td><content ID="d5">diffus</content></td>
| |
| − | <td><content ID="d6">Formalin</content></td>
| |
| − | <td><content ID="d7">Tumor isolierte Tumorzelle</content></td>
| |
| − | </tr>
| |
| − | ...
| |
| − | </tbody>
| |
| − | </text>
| |
| − | | |
| − | <!-— erste Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="????"
| |
| − | codeSystem="??????"
| |
| − | displayName="Antikörperfärbung Art (kurz)" />
| |
| − | <value xsi:type="CD" code="?????" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d1'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— zweite Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation>
| |
| − | <code code="????"
| |
| − | codeSystem="??????"
| |
| − | displayName="Antikörperfärbung Reaktion" />
| |
| − | <value xsi:type="CD" code="2" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d2'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— dritte Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="xxxx"
| |
| − | codeSystem="a.b.c.dx.y.z"
| |
| − | displayName="Antikörperfärbung Reaktionsstärke" />
| |
| − | <value xsi:type="CD" code="3" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d3'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— vierte Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="xxxx"
| |
| − | codeSystem="a.b.c.dx.y.z"
| |
| − | displayName="Antikörperfärbung Prozent" />
| |
| − | <value xsi:type="CD" code="3" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d4'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— fünfte Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="xxxx"
| |
| − | codeSystem="a.b.c.dx.y.z"
| |
| − | displayName="Verteilung" />
| |
| − | <value xsi:type="CD" code="3" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d5'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— sechste Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="xxxx"
| |
| − | codeSystem="a.b.c.dx.y.z"
| |
| − | displayName="Fixierung" />
| |
| − | <value xsi:type="CD" code="3" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d6"'''</u>/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— siebte Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="xxxx"
| |
| − | codeSystem="a.b.c.dx.y.z"
| |
| − | displayName="Gewebe" />
| |
| − | <value xsi:type="CD" code="3" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d7'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | ...
| |
| − | | |
| − | </section>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | Definierte Vokabeldomänen:
| |
| − | * 4.3.1. Antikörper
| |
| − | * 4.3.2. Färbung
| |
| − | * 4.3.3. Fixierung
| |
| − | * 4.3.4. Reaktion
| |
| − | * 4.3.5. Verteilung
| |
| − | * 4.3.6. Gewebe
| |
| − | | |
| − | =====Beispiel=====
| |
| − | | |
| − | {{BeginPurpleBox|Beispiel}}
| |
| − | PathoBerichtText
| |
| − | | |
| − | Wiederholung der Ki-67-Färbung von der 2. Fraktion (rechts).
| |
| − | | |
| − | Beurteilung
| |
| − | | |
| − | 1. Isolierte Tumorzellen in einem funktionsgesteigerten Lymphknoten (linke Axilla Sentinellymphknoten 544cps). | |
| − | | |
| − | 2. Teils glanduläres, teils solides, invasives ductales Adenocarcinom der Brustdrüse, geringe nukleäre Atypien sowie ductales Carcinoma in situ mit geringen Atypien, DCIS I und Mikrokalk bis 0,2mm (rechte Mamma oben zwischen den Quadranten, Resektat); immunhistologisch fokal starke Expression des Östrogenrezeptors in etwa 60% der Tumorzellen.
| |
| − | Score nach Elston und Ellis: 4.
| |
| − | Immunreaktiver Score: Östrogenrezeptor 9, Progesteronrezeptor 0.
| |
| − | Onkoprotein C-erbB-2 Index: 0<br>Größter Durchmesser des invasiven Carcinoms etwa 9mm, das invasive Carcinom reicht zumindest an die craniale Abtragungsebene im peripheren (cranialen) Pol. Durchmesser der in situ Komponente etwa 35mm, Abstand von der nächstgelegenen (posterioren) Abtragungsebene etwa 0,4mm, die in situ Komponente breitet sich allerdings ebenfalls in den peripheren (cranialen) Pol aus und erreicht damit dort mindestens die Abtragungsebene.
| |
| − | | |
| − | 3. Weitgehend solides, invasives ductales Adenocarcinom der Brustdrüse, mittelgradige, herdförmig schwere nukleäre Atypien, etwa in gleicher Größe ductales Carcinoma in situ mit schweren Atypien, DCIS III sowie Nekrosen und Verkalkungen bis 4mm (Resektat linke Mamma oben außen); immunhistologisch starke Expression des Östrogenrezeptors und des Progesteronrezeptors jeweils in etwa 85% der Tumorzellen.
| |
| − | Immunreaktiver Score: Östrogenrezeptor 12, Progesteronrezeptor 12.
| |
| − | Kleine Wachstumsfraktion (Ki-67 um 10 %).
| |
| − | Onkoprotein C-erbB-2 (Her-2-Neu-Index: 0). Größter Durchmesser der invasiven Komponente und der in-situ-Komponente jeweils etwa 23mm. Abstand der invasiven Komponente von der nächstgelegenen Abtragungsebene (posterior) 3,2mm, Abstand von anterior 12mm, von cranial 20mm, von caudal 23mm, von medial 30mm, von lateral 13mm. Die in-situ-Komponente breitet sich zumindest bis an die posteriore Abtragungsebene aus.
| |
| − | | |
| − | 4. Neun tumorfreie Lymphknoten (linke Axilla).
| |
| − | | |
| − | 5. Tumorfreies Fettgewebe (linke Mamma, Nachresektat lateral).
| |
| − | | |
| − | 6. Überwiegend tumorfreies Fettgewebe mit kleinen Anteilen von tumorfreiem Brustdrüsengewebe (linke Mamma, Nachresektat mamillenwärts).
| |
| − | | |
| − | 7. Überwiegend tumorfreies Fettgewebe mit kleinen Anteilen von tumorfreiem Brustdrüsengewebe (linke Mamma, Nachresektat cranial).
| |
| − | | |
| − | Auch nach Wiederholung lässt sich die Wachstumsfraktion in dem kleinen Tumor auf der rechten Seite nicht darstellen, vermutlich wurde das Antigen durch die schwere thermische Schädigung zerstört.
| |
| − | | |
| − | {{EndPurpleBox}}
| |
| − | | |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <StructuredBody>
| |
| − | <component>
| |
| − | <section>
| |
| − | <code code="PathoBerichtText" codeSystem="1.2.276.0.76.5.??????"/>
| |
| − | <title></title>
| |
| − | <text>
| |
| − | Wiederholung der Ki-67-Färbung von der 2. Fraktion (rechts).
| |
| − | </text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component>
| |
| − | <section>
| |
| − | <code code="PathoBerichtText" codeSystem="1.2.276.0.76.5.??????"/>
| |
| − | <title>Beurteilung</title>
| |
| − | <text>
| |
| − | 1. Isolierte Tumorzellen in einem funktionsgesteigerten Lymphknoten
| |
| − | (linke Axilla Sentinellymphknoten 544cps).<br>
| |
| − | <br>
| |
| − | 2. Teils glanduläres, teils solides, invasives ductales Adenocarcinom der Brustdrüse,
| |
| − | geringe nukleäre Atypien sowie ductales Carcinoma in situ mit geringen Atypien, DCIS I
| |
| − | und Mikrokalk bis 0,2mm (rechte Mamma oben zwischen den Quadranten, Resektat);
| |
| − | immunhistologisch fokal starke Expression des Östrogenrezeptors in etwa 60% der
| |
| − | Tumorzellen. <br>
| |
| − | Score nach Elston und Ellis: 4.<br>
| |
| − | Immunreaktiver Score: Östrogenrezeptor 9, Progesteronrezeptor 0.<br>
| |
| − | Onkoprotein C-erbB-2 Index: 0<br>Größter Durchmesser des invasiven Carcinoms etwa 9mm,
| |
| − | das invasive Carcinom reicht zumindest an die craniale Abtragungsebene im peripheren
| |
| − | (cranialen) Pol. Durchmesser der in situ Komponente etwa 35mm, Abstand von der
| |
| − | nächstgelegenen (posterioren) Abtragungsebene etwa 0,4mm, die in situ Komponente breitet
| |
| − | sich allerdings ebenfalls in den peripheren (cranialen) Pol aus und erreicht damit dort
| |
| − | mindestens die Abtragungsebene.<br>
| |
| − | <br>
| |
| − | 3. Weitgehend solides, invasives ductales Adenocarcinom der Brustdrüse, mittelgradige,
| |
| − | herdförmig schwere nukleäre Atypien, etwa in gleicher Größe ductales Carcinoma in situ
| |
| − | mit schweren Atypien, DCIS III sowie Nekrosen und Verkalkungen bis 4mm (Resektat linke
| |
| − | Mamma oben außen); immunhistologisch starke Expression des Östrogenrezeptors und des
| |
| − | Progesteronrezeptors jeweils in etwa 85% der Tumorzellen.<br>
| |
| − | Immunreaktiver Score: Östrogenrezeptor 12, Progesteronrezeptor 12. <br>
| |
| − | Kleine Wachstumsfraktion (Ki-67 um 10 %). <br>
| |
| − | Onkoprotein C-erbB-2 (Her-2-Neu-Index: 0). Größter Durchmesser der invasiven Komponente
| |
| − | und der in-situ-Komponente jeweils etwa 23mm. Abstand der invasiven Komponente von der
| |
| − | nächstgelegenen Abtragungsebene (posterior) 3,2mm, Abstand von anterior 12mm, von cranial
| |
| − | 20mm, von caudal 23mm, von medial 30mm, von lateral 13mm. Die in-situ-Komponente breitet
| |
| − | sich zumindest bis an die posteriore Abtragungsebene aus. <br>
| |
| − | <br>
| |
| − | 4. Neun tumorfreie Lymphknoten (linke Axilla).<br>
| |
| − | <br>
| |
| − | 5. Tumorfreies Fettgewebe (linke Mamma, Nachresektat lateral).<br>
| |
| − | <br>
| |
| − | 6. Überwiegend tumorfreies Fettgewebe mit kleinen Anteilen von tumorfreiem
| |
| − | Brustdrüsengewebe (linke Mamma, Nachresektat mamillenwärts).<br>
| |
| − | <br>
| |
| − | 7. Überwiegend tumorfreies Fettgewebe mit kleinen Anteilen von tumorfreiem
| |
| − | Brustdrüsengewebe (linke Mamma, Nachresektat cranial).
| |
| − | </text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component>
| |
| − | <section>
| |
| − | <code code="PathoBerichtText" codeSystem="1.2.276.0.76.5.??????"/>
| |
| − | <title></title>
| |
| − | <text>
| |
| − | Auch nach Wiederholung lässt sich die Wachstumsfraktion in dem kleinen Tumor auf der
| |
| − | rechten Seite nicht darstellen, vermutlich wurde das Antigen durch die schwere thermische
| |
| − | Schädigung zerstört.
| |
| − | </text>
| |
| − | </section>
| |
| − | </component>
| |
| − | </StructuredBody>
| |
| − | </syntaxhighlight>
| |
| − | | |
| − | ====Attribut-Wert-Paare====
| |
| − | Die Attribut-Wert-Paare werden textuell aus den codierten Informationen abgeleitet (derived).
| |
| − | Die zu verwendenden Vokabularien sind im Anhang detailliert aufgelistet.
| |
| − | | |
| − | Die Informationen werden als Attribut-Wert-Paare in Form einer Tabelle dargestellt, die wie folgt aussieht.
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Entnahme!!Resektat
| |
| − | |-
| |
| − | |Kalk histologisch||Ja
| |
| − | |-
| |
| − | |Kalk (mm)||0,2
| |
| − | |-
| |
| − | |}
| |
| − | | |
| − | Oder in XML:
| |
| − | <syntaxhighlight lang="xml">
| |
| − | <section>
| |
| − | <!-- Darstellung als Tabelle -->
| |
| − | <text>
| |
| − | <tbody>
| |
| − | <tr>
| |
| − | <td><content ID="'''<u>d1'''</u>">Entnahme</content></td>
| |
| − | <td>Resektat</td>
| |
| − | </tr>
| |
| − | <tr>
| |
| − | <td><content ID="'''<u>d2'''</u>">Kalk Histologisch</content></td>
| |
| − | <td>Ja</td>
| |
| − | </tr>
| |
| − | <tr>
| |
| − | <td><content ID="'''<u>d3'''</u>">Kalk (mm)</content></td>
| |
| − | <td>0,2</td>
| |
| − | </tr>
| |
| − | ...
| |
| − | </tbody>
| |
| − | </text>
| |
| − | | |
| − | <!-— erste Information -->
| |
| − | <entry typeCode="DRIV">>
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code"Mamma.Entnahme" codeSystem="??????" />
| |
| − | <value xsi:type="CD" code="Resektat" codeSystem="????">
| |
| − | <originalText><reference value="\#'''<u>d1'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— zweite Information -->
| |
| − | <entry typeCode="DRIV">
| |
| − | <observation>
| |
| − | <code codeMamma.Kalk Histologisch" codeSystem="??????" />
| |
| − | < value xsi:type="BL" code="true">
| |
| − | <originalText><reference value="\#'''<u>d2'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!-— dritte Information -->
| |
| − | <entry typeCode="DRIV">>
| |
| − | <observation classCode="OBS" moodCode="EVN"> | |
| − | <code code="Mamma.Kalk"
| |
| − | codeSystem="a.b.c.dx.y.z"
| |
| − | displayName="Kalk" />
| |
| − | <value xsi:type="PQ" value="0,2" unit="mm" >
| |
| − | <originalText><reference value="\#'''<u>d3'''</u>"/></originalText>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | | |
| − | <!—weitere Information -->
| |
| − | ...
| |
| − | </section>
| |
| − | </syntaxhighlight>
| |
| − | | |
| − | ====40. kritische gutachterliche Stellungnahme====
| |
| − | | |
| − | | |
| − | ====41. Beurteilung====
| |
| − | | |
| − | | |
| − | ====42. Kommentar====
| |
| − | | |
| − | | |
| − | ====43. Weitergabemodus====
| |
| − | | |
| − | | |
| − | ====44. Gruß====
| |
| − | Dies ist bereits Bestandteil des VHitG-Arztbriefes.
| |
| − | | |
| − | =45. Vokabeldomänen=
| |
| − | | |
| − | | |
| − |
| |
| − | | |
| − | | |
| − | ==45. Einleitung==
| |
| − | Dieser Abschnitt dient der Trennung von verwendeten Codes und der normativen Spezifikation. Damit lassen sich die Codes aktualisieren, ohne dass die Spezifikation überarbeitet werden muss.
| |
| − | | |
| − | Dieser Abschnitt ist deshalb nur informativ. Die jeweils aktuellen Codes sind deshalb zu erfragen.
| |
| − | | |
| − | ==46. Überblick über die Codierschemata==
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Vokabeldomäne/ Codiersystem!!OID!!Kurzbezeichnung!!Diagnosen!!Lokalisationen
| |
| − | |-
| |
| − | |ICD10GM||||||||
| |
| − | |-
| |
| − | |ICD-10 GM Version 2010||1.2.276.0.76.5.384||icd10gm2010||x||
| |
| − | |-
| |
| − | |ICD-10 GM Version 2009||1.2.276.0.76.5.356||icd10gm2009||x||
| |
| − | |-
| |
| − | |ICD-10 GM Version 2008||1.2.276.0.76.5.330||icd10gm2008||x||
| |
| − | |-
| |
| − | |ICD-10 GM Version 2007||1.2.276.0.76.5.318||icd10gm2007||x||
| |
| − | |-
| |
| − | |ICD-10 GM Version 2006||1.2.276.0.76.5.311||icd10gm2006||x||
| |
| − | |-
| |
| − | |ICD-O||||||||
| |
| − | |-
| |
| − | |ICD-O-3||||icd-o-3||||
| |
| − | |-
| |
| − | |ICD-O-DA-1978||||||||
| |
| − | |-
| |
| − | |ICD-O-DA-2002||||||||
| |
| − | |-
| |
| − | |TNM||||||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.341||c-faktor-tumor||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.340||tnm-qualifier||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.339||metastasen||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.338||nodus-tnm||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.337||ausdehnung-tnm||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.336||diff-grading-tumor||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.335||dignitaet-tumor||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.334||tumordiagnosen||||
| |
| − | |-
| |
| − | |Alpha-ID||||||||
| |
| − | |-
| |
| − | |Alpha-ID 2010||1.2.276.0.76.5.383||alphaid2010||x||
| |
| − | |-
| |
| − | |Alpha-ID 2009||1.2.276.0.76.5.355||alphaid2009||x||
| |
| − | |-
| |
| − | |Alpha-ID 2008||1.2.276.0.76.5.329||alphaid2008||x||
| |
| − | |-
| |
| − | |Alpha-ID 2007||1.2.276.0.76.5.316||alphaid2007||x||
| |
| − | |-
| |
| − | |Alpha-ID 2006||1.2.276.0.76.5.309||alphaid2006||x||
| |
| − | |-
| |
| − | |MeSH||||||||
| |
| − | |-
| |
| − | |MeSH||2.16.840.1.113883.6.177.5||MSHGER||x||x
| |
| − | |-
| |
| − | |Kodiersysteme||||||||
| |
| − | |-
| |
| − | |Snomed CT||2.16.840.1.113883.6.96||SNOMED CT||x||x
| |
| − | |-
| |
| − | |ID Macs||1.2.276.0.76.5.305||id_macs||x||x
| |
| − | |-
| |
| − | |LOINC||2.16.840.1.113883.6.1||loinc||x||
| |
| − | |-
| |
| − | |||||||||
| |
| − | |-
| |
| − | |||1.2.276.0.76.5.342||typisierung-diagnose||x||x
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 5: Codierschemata
| |
| − | | |
| − | ==Codes für Färbungen==
| |
| − | | |
| − | ===Antikörper===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Antikörper!!Bedeutung
| |
| − | |-
| |
| − | |..||||
| |
| − | |-
| |
| − | |..||||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 6: Antikörper (OID: (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===Färbung===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |0||negativ||
| |
| − | |-
| |
| − | |1||fraglich positiv||
| |
| − | |-
| |
| − | |2||positiv||
| |
| − | |-
| |
| − | |9||nicht auswertbar||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 7: Färbung (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===50. Fixierung===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |..||Formalin||
| |
| − | |-
| |
| − | |||||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 8: Fixierung (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===Reaktion===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |0||keine||
| |
| − | |-
| |
| − | |1||schwach||
| |
| − | |-
| |
| − | |2||mittel||
| |
| − | |-
| |
| − | |3||stark||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 9: Reaktion (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===Verteilung===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |0||||
| |
| − | |-
| |
| − | |1||diffus||
| |
| − | |-
| |
| − | |2||fokal||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 10: Verteilung (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===Gewebe===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Gewebe!!Bedeutung
| |
| − | |-
| |
| − | |||Turmor||
| |
| − | |-
| |
| − | |||Tumor (isolierte Zelle)||
| |
| − | |-
| |
| − | |||Kontrollgewebe||
| |
| − | |-
| |
| − | |||DCIS||
| |
| − | |-
| |
| − | |||Lymphknoten||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 11: Gewebe(OID: (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ==54. spezielle Codes für Attribut-Wert-Paare==
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung!!Datentyp!!Einheiten
| |
| − | |-
| |
| − | |||Abtragungsebene klinisch markiert||||BL||
| |
| − | |-
| |
| − | |||Amputation||||BL||
| |
| − | |-
| |
| − | |||Aneurysma||||BL||
| |
| − | |-
| |
| − | |||Aneurysma dissecans||||BL||
| |
| − | |-
| |
| − | |||Aneurysma spurium||||BL||
| |
| − | |-
| |
| − | |||Angiodysplasie||||BL||
| |
| − | |-
| |
| − | |||Aplasie||||BL||
| |
| − | |-
| |
| − | |||Appendix Torsion||||BL||
| |
| − | |-
| |
| − | |||Artherie Trombus||||BL||
| |
| − | |-
| |
| − | |||Arteriosclerose||||BL||
| |
| − | |-
| |
| − | |||Arteriovenöse Malformation||||BL||
| |
| − | |-
| |
| − | |||Asbest||||BL||
| |
| − | |-
| |
| − | |||Asbest Exposition klinisch||||BL||
| |
| − | |-
| |
| − | |||Asbestose klinisch||||BL||
| |
| − | |-
| |
| − | |||Atypische epitheliale Proliferation ductal||||BL||
| |
| − | |-
| |
| − | |||Bezoar||||BL||
| |
| − | |-
| |
| − | |||Kalk, histologisch||||BL||
| |
| − | |-
| |
| − | |||Kalk||||PQ||mm
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 12: Attribut-Wert-Paare (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===55. Entnahme===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |||Abort||
| |
| − | |-
| |
| − | |||Abradat||
| |
| − | |-
| |
| − | |||Abradat klinisch post Abortum||
| |
| − | |-
| |
| − | |||Abradat klinisch post Partum||
| |
| − | |-
| |
| − | |||Abstrich||
| |
| − | |-
| |
| − | |||Amputation||
| |
| − | |-
| |
| − | |||Amputation quartaer||
| |
| − | |-
| |
| − | |||Amputation sekundaer||
| |
| − | |-
| |
| − | |||Amputation tertiaer||
| |
| − | |-
| |
| − | |||Aquadissektion||
| |
| − | |-
| |
| − | |||...||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 13: Entnahme (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===56. Beurteilbarkeit===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |||gut||
| |
| − | |-
| |
| − | |||ausreichend||
| |
| − | |-
| |
| − | |||eingeschraenkt||
| |
| − | |-
| |
| − | |||unzureichend||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 14: Beurteilbarkeit (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===57. Abort===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |||unklar||
| |
| − | |-
| |
| − | |||Uterin||
| |
| − | |-
| |
| − | |||Extrauterin||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 15: Abort (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===58. Adenose===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |||sklerosierend||
| |
| − | |-
| |
| − | |||sklerosierend nodular||
| |
| − | |-
| |
| − | |||Blunt duct||
| |
| − | |-
| |
| − | |||microglandular||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 16: Adenose (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===59. Anastomose===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |||regelhaft||
| |
| − | |-
| |
| − | |||Ulcus||
| |
| − | |-
| |
| − | |||turmorrezidiv||
| |
| − | |-
| |
| − | |||Entzündung unklassifiziert||
| |
| − | |-
| |
| − | |||Insuffizienz klinisch||
| |
| − | |-
| |
| − | |||Insuffizient möglich||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 17: Anastomose (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===60. Atrophie===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |||gering||
| |
| − | |-
| |
| − | |||mittelgradig||
| |
| − | |-
| |
| − | |||schwergradig||
| |
| − | |-
| |
| − | |||gering fokal||
| |
| − | |-
| |
| − | |||mittelgradig fokal||
| |
| − | |-
| |
| − | |||schwergradig fokal||
| |
| − | |-
| |
| − | |||gering partiell||
| |
| − | |-
| |
| − | |||mittelgradig partiell||
| |
| − | |-
| |
| − | |||schwergradig partiell||
| |
| − | |-
| |
| − | |||gering diffus||
| |
| − | |-
| |
| − | |||mittelgradig diffus||
| |
| − | |-
| |
| − | |||schwergradig diffus||
| |
| − | |-
| |
| − | |||nein||
| |
| − | |-
| |
| − | |||gering überwieged||
| |
| − | |-
| |
| − | |||mittelgradig überwiegend||
| |
| − | |-
| |
| − | |||schwergradig überwiegend||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 18: Atrophie (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | {{AlertBox|
| |
| − | Sollen die Tabellen so aufbereitet werden, wie vorhergehend dargestellt?<br>
| |
| − | Oder sollen wir einen generischen Mechanismus verwenden, so wie in der folgenden Auflistung. }}
| |
| − | | |
| − | ==61. generische Codes für Attribut-Wert-Paare==
| |
| − | | |
| − | | |
| − | ===62. Interpretation===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | | ||negativ||
| |
| − | |-
| |
| − | | ||positiv||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 19: Interpretation (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | Hier stellt sich die Frage, ob bestimmte Attribute nicht auch über boolesche Werte abgebildet werden können?
| |
| − | | |
| − | ==63. boolesche Attribute==
| |
| − | Die folgende Tabelle listet die Attribute, die boolesche Werte ausgedrückt werden können:
| |
| − | {| class="hl7table"
| |
| − | !Attribut!!ja!!nein!!möglich!!unklar!!unbekannt!!Verdacht!!nicht untersucht!!nicht beurteilbar!!fraglich
| |
| − | |-
| |
| − | |Abtragungsebene Klinik markiert||x||x||||||||||||||
| |
| − | |-
| |
| − | |Aneurysma dissecans||x||||||||||||||||
| |
| − | |-
| |
| − | |Aneurysma spurium||x||||x||||||||||||
| |
| − | |-
| |
| − | |Angiodysplasie||x||x||x||||||||||||
| |
| − | |-
| |
| − | |Aplasie||||||x||||||||||||
| |
| − | |-
| |
| − | |Appendix Torsion||||||x||||||||||||
| |
| − | |-
| |
| − | |Arterie Thrombus||x||||||||||||||||
| |
| − | |-
| |
| − | |Asbest||x||x||||||||||||||
| |
| − | |-
| |
| − | |Asbest Exposition klinisch||x||||||||x||||||||
| |
| − | |-
| |
| − | |Asbestose klinisch||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Atypische epitheliale Proliferation ductal||x||x||||||||||||||
| |
| − | |-
| |
| − | |Atypische epitheliale Proliferation in Papillom||x||x||||||||||||||
| |
| − | |-
| |
| − | |Atypische vaskulaere Laesion||||x||x||||||||||||
| |
| − | |-
| |
| − | |Ausbreitung im Nierenbecken||x||x||||||||||x||x||
| |
| − | |-
| |
| − | |Ausbreitung in Nebenniere||x||x||||||||||x||||
| |
| − | |-
| |
| − | |Ausbreitung perihilaer||x||x||||||||||||||
| |
| − | |-
| |
| − | |Ausbreitung perirenal||x||x||||||||||||||
| |
| − | |-
| |
| − | |Ausbreitung Rete||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Ausbreitung Tunica albuginea||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Bandscheibe Degeneration||x||x||||||||||||||
| |
| − | |-
| |
| − | |Bandscheibe Ruptur||x||||x||||||||||||
| |
| − | |-
| |
| − | |Barrett-Mucosa klinisch||x||x||||||x||x||||||
| |
| − | |-
| |
| − | |Barrett-Mukosa||x||x||x||||||||x||||
| |
| − | |-
| |
| − | |Basis Klinik markiert||x||x||||||||||||||
| |
| − | |-
| |
| − | |Bezoar||x||||||||||||||||
| |
| − | |-
| |
| − | |Bezoar klinisch||x||||||||||||||||
| |
| − | |-
| |
| − | |Biopsat kontralateral falsch negativ||x||||||||||||||||
| |
| − | |-
| |
| − | |Bypass klinisch||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Calcinose||x||||||||||||||||
| |
| − | |-
| |
| − | |Chorangiom||x||||||||||||||||
| |
| − | |-
| |
| − | |Chorangiose||x||x||x||||||||||||
| |
| − | |-
| |
| − | |Colitis ulcerosa DALM||x||x||||||||||||||
| |
| − | |-
| |
| − | |Colitis ulcerosa DALM-Referenzgutachten||x||x||||||||||||||
| |
| − | |-
| |
| − | |DCIS extensiv (4:1)||x||x||||||||||||||
| |
| − | |-
| |
| − | |Decidua||x||x||||||||||||||
| |
| − | |-
| |
| − | |Divertikel||||||x||||||||||||
| |
| − | |-
| |
| − | |Divertikel klinisch||x||||||||||||||||
| |
| − | |-
| |
| − | |Döderleinflora||x||x||||||||||||||
| |
| − | |-
| |
| − | |Druesenkoerperzysten||x||||||||||||||||
| |
| − | |-
| |
| − | |Ductus deferens vollstaendig circulaer||x||||||||||||||||
| |
| − | |-
| |
| − | |Duodenalwandzyste||x||||||||||||||||
| |
| − | |-
| |
| − | |Dysplasie||x||||x||||||||||||
| |
| − | |-
| |
| − | |Dystopie||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Eisenkoerper||x||x||||||||||||||
| |
| − | |-
| |
| − | |Endangiopathia obliterans||x||||x||||||||||||
| |
| − | |-
| |
| − | |Endozervikale Zellen||x||x||||||||||||||
| |
| − | |-
| |
| − | |Eosinophilie||x||x||||||||||||||
| |
| − | |-
| |
| − | |Epithelproliferation||x||||||||||||||||
| |
| − | |-
| |
| − | |Familiaeres Risiko||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Fetofetale Transfusion||||||x||||||||||||
| |
| − | |-
| |
| − | |Fettgewebe Hyperplasie nodulaer||x||||||||||||||||
| |
| − | |-
| |
| − | |Fibroadenom||x||||x||||||||||||
| |
| − | |-
| |
| − | |Fibroadenomatoide Mastopathie||x||||||||||x||||||
| |
| − | |-
| |
| − | |Fissur||||||x||x||||||||||
| |
| − | |-
| |
| − | |Fistel||x||||||||||||||||
| |
| − | |-
| |
| − | |Flache epitheliale Atypie||x||||||||||||||||
| |
| − | |-
| |
| − | |Fraktur Nekrose||||||x||||||||||||
| |
| − | |-
| |
| − | |Fremdkoerper||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Fremdkoerper Silikon||||x||x||||||||||||
| |
| − | |-
| |
| − | |Funiculocele||||||||||||||||||
| |
| − | |-
| |
| − | |Funiculocele||||||x||||||||||||
| |
| − | |-
| |
| − | |Gastrale antrale vaskulaere Ektasie||||||x||||||||||||
| |
| − | |-
| |
| − | |Gastrale antrale vaskulaere Ektasie||||x||||||||||||||
| |
| − | |-
| |
| − | |GERD||||||x||||||||||||
| |
| − | |-
| |
| − | |Gewebe enthalten||||x||||||||||||||
| |
| − | |-
| |
| − | |Graviditaet klinisch||x||||||||x||||||||
| |
| − | |-
| |
| − | |Gynaekomastie||x||||x||||||||||||
| |
| − | |-
| |
| − | |Haematom retroplazentar||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Haemorrhoide||||||x||||||||||||
| |
| − | |-
| |
| − | |Hamartom||||||x||||||||||||
| |
| − | |-
| |
| − | |Haut Tumorinfiltration||x||x||||||||||||||
| |
| − | |-
| |
| − | |Haut ulceriert||x||x||||||||||||||
| |
| − | |-
| |
| − | |Hautnarbe||x||||||||||||||||
| |
| − | |-
| |
| − | |Hautnarbe Keloid||x||||||||||||||||
| |
| − | |-
| |
| − | |Helicobacter||x||x||||||||||x||||
| |
| − | |-
| |
| − | |Hormone||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Hydatide||||||x||||||||||||
| |
| − | |-
| |
| − | |Hydrocele||x||||x||||||||||||
| |
| − | |-
| |
| − | |Hydrosalpinx||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Hyperplasie C-Zelle||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Hyperplasie ductal Atypie (ADH)||x||||||||||||||||
| |
| − | |-
| |
| − | |Hyperplasie ductal einfach||x||x||||||||||||||
| |
| − | |-
| |
| − | |Hyperplasie endocrin||x||||||||||||||||
| |
| − | |-
| |
| − | |Hyperplasie pseudoangiomatoes stromal||x||||||||||||||||
| |
| − | |-
| |
| − | |Indikation||||||||||x||||||||
| |
| − | |-
| |
| − | |Infarkt||x||x||||||||||||||
| |
| − | |-
| |
| − | |Insuffizienz||x||||x||||||||||||
| |
| − | |-
| |
| − | |Intraduktaler Anteil||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk Assoziation||||||||x||||||||||
| |
| − | |-
| |
| − | |Kalk benigne Laesion||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk benigne und maligne Laesion||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk histologisch||x||x||||||||||||||
| |
| − | |-
| |
| − | |Kalk histologisch ADH||x||x||||||||||||||
| |
| − | |-
| |
| − | |Kalk histologisch CIS||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk histologisch DCIS||x||x||||||||||||||
| |
| − | |-
| |
| − | |Kalk histologisch FEA||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk histologisch LCIS||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk maligne Laesion||x||||||x||||||||||
| |
| − | |-
| |
| − | |Kalk Oxalat||x||||||||||||||||
| |
| − | |-
| |
| − | |Kalk radiologisch||x||x||||x||x||||||||x
| |
| − | |-
| |
| − | |Kalk radiologisch Groesse||||||||||x||||||||
| |
| − | |-
| |
| − | |Kalk unklare Laesion||x||||||||||||||||
| |
| − | |-
| |
| − | |Keratose aktinisch||||||x||||||||||||
| |
| − | |-
| |
| − | |Klin Fazialisresektion||||x||||||||||||||
| |
| − | |-
| |
| − | |Klin Malignitätsverdacht||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klin Resektion extrakapsulaer||x||||||||||||||||
| |
| − | |-
| |
| − | |Klin Resektion vollstaendig||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klin Sialoadenektomie partiell||x||||||||||||||||
| |
| − | |-
| |
| − | |Klin Sialoadenektomie total||x||||||||||||||||
| |
| − | |-
| |
| − | |Klin Tumor Durchmesser||||||||||x||||||||
| |
| − | |-
| |
| − | |Klin Tumor intakt||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinik Abtragungsebene markiert||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinik Levator markiert||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinik Lipom Hernie||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinik Otosklerose||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinik Polyp||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinik R-Klassification||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinik Zyklustermin angegeben||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Amnioninfektionssyndrom||x||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Anastomose||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Angiodysplasie||x||||x||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Aplasie||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Blutung postpartal||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Bowenoide Papulose||||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Calciphylaxie||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Clipping||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Conn-Syndrom||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch CTG pathologisch||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Descensus||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Diabetes||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Diabetes mellitus||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Divertikel Meckel||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Ductus omphaloentericus||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Dysgenesie||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Dysplasie||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Ehlers-Danlos||||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Ektopie Verdacht||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinisch endokrin aktiv||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch EPH-Gestose||x||||x||||||||||||
| |
| − | |-
| |
| − | |Klinisch Eradikation||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Erosion||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch fetofetale Transfusion||||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Fraktur||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Fruchtwasser gruen||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Gastrostoma||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Gonadendysgenesie||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Gonadendysgenesie XY||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Haematom retroplazentar||x||x||x||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Haemorrhoide||x||||x||||||||||||
| |
| − | |-
| |
| − | |Klinisch HELLP-Syndrom||x||||x||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Hydatide||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Hyperaldosteronismus||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Hypertonus renal||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Ileus||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Inkarzeration||x||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Insuffizienz||x||||x||||x||x||||||
| |
| − | |-
| |
| − | |Klinisch intracraniell||x||x||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Kalter Knoten||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Karpaltunnelsyndrom||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Loesung vorzeitig||x||||||||x||x||||||
| |
| − | |-
| |
| − | |Klinisch Meniscusruptur||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Nabelschnurknoten||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Nabelschnurumschlingung||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Neurogene Entleerungsstoerung||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Operationswunde||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Paraphimose||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Perforation||x||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Phimose||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Phimose absolut||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Phimose relativ||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Plazenta accreta||x||x||||||x||x||||||
| |
| − | |-
| |
| − | |Klinisch Plazenta increta||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Polgefaess||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Portale Hypertensive Gastropathie||x||||||||x||x||||||
| |
| − | |-
| |
| − | |Klinisch Prolaps||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Prolaps Recidiv||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Pseudarthrose||||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Radioderm||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Reflux||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Reinke Oedem||x||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Revaskularisation||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Ruptur||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Ruptur Trauma||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Sigma elongatum||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch solitaere Nabelschnurarterie||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Sterilisation||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Transsexualitaet||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Trauma||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Trauma Ruptur||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Trauma Verbrennung||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Ulcus||x||x||x||||x||||||||
| |
| − | |-
| |
| − | |Klinisch Ureter Ruptur Trauma iatrogen||x||||||||||||||||
| |
| − | |-
| |
| − | |Klinisch Varicocele||x||||||||x||||||||
| |
| − | |-
| |
| − | |Klinisch velamentoeser Nabelschnuransatz||x||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch vorzeitiger Blasensprung||x||||||||||x||||||
| |
| − | |-
| |
| − | |Klinisch Zyste||x||x||||||x||x||||||
| |
| − | |-
| |
| − | |Knorpel||x||||||||||||||||
| |
| − | |-
| |
| − | |Koilozyten||x||x||||||||||||||
| |
| − | |-
| |
| − | |Kokkenflora||x||x||||||||||||||
| |
| − | |-
| |
| − | |Kollagen mikronodulaer||x||||||||||||||||
| |
| − | |-
| |
| − | |Kollagene Sphaerulose||x||||||||||||||||
| |
| − | |-
| |
| − | |Komplexe sklerosierende Laesion||x||||x||||||||||||
| |
| − | |-
| |
| − | |Konkremente||x||x||||||||||||||
| |
| − | |-
| |
| − | |Korrelat zur Klinik||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Korrelation mit Klinik||x||||||x||||||||||
| |
| − | |-
| |
| − | |Korrelation mit Radiologie||x||||||x||||||||||
| |
| − | |-
| |
| − | |Korrelation mit Stanzbiopsie||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Kreislaufstoerung Infarkt||||||x||||||||||||
| |
| − | |-
| |
| − | |Kreislaustoerung Infarzierung||||||x||||||||||||
| |
| − | |-
| |
| − | |Lipidinsel||x||||||||||||||||
| |
| − | |-
| |
| − | |Lithiasis||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Lithiasis klinisch||x||||||||x||||||||
| |
| − | |-
| |
| − | |Loesung vorzeitig||x||||x||||||||||||
| |
| − | |-
| |
| − | |Lokalisation||||||||||x||||||||
| |
| − | |-
| |
| − | |Lymphknoten||x||||||||||||||||
| |
| − | |-
| |
| − | |Lymphknoten intramammaer||x||||||||||||||||
| |
| − | |-
| |
| − | |Medullaeres Karzinom klinisch Verdacht||x||x||||||x||||||||
| |
| − | |-
| |
| − | |Mekoniumphagozytose||x||x||||||||||||||
| |
| − | |-
| |
| − | |Meniscus Degeneration||x||||x||||||||||||
| |
| − | |-
| |
| − | |Meniscusruptur||x||||x||||||||||||
| |
| − | |-
| |
| − | |Meniscusruptur Recidiv||x||||||||||||||||
| |
| − | |-
| |
| − | |Mesangiosclerose||x||||||||||||||||
| |
| − | |-
| |
| − | |Mesangiosclerose nodulaer||x||||||||||||||||
| |
| − | |-
| |
| − | |Mesonephrogen Rest||x||||||||||||||||
| |
| − | |-
| |
| − | |Mucocele||x||x||x||||||||||||
| |
| − | |-
| |
| − | |MucosaprolapsSyndrom moeglich||x||||||||||||||||
| |
| − | |-
| |
| − | |Muskulatur basal||x||x||||||||||||||
| |
| − | |-
| |
| − | |Mutation k-ras||x||x||||||||||||||
| |
| − | |-
| |
| − | |Nabelschnurknoten||x||||||||||||||||
| |
| − | |-
| |
| − | |Pigmentierung||x||||||||||||||||
| |
| − | |-
| |
| − | |Pilze||x||x||||||||||||||
| |
| − | |-
| |
| − | |Plazenta accreta||x||x||x||||||||||||
| |
| − | |-
| |
| − | |Plazenta bilobata||x||||||||||||||||
| |
| − | |-
| |
| − | |Plazenta increta||||||x||||||||||||
| |
| − | |-
| |
| − | |Plazenta Trophoblastreaktion ueberschiessend||||||x||||||||||||
| |
| − | |-
| |
| − | |Pneumatisationskammer||x||||||||||||||||
| |
| − | |-
| |
| − | |Pneumatosis coli||||||x||||||||||||
| |
| − | |-
| |
| − | |Polyp fibroepithelial||x||||||||||||||||
| |
| − | |-
| |
| − | |Portale Hypertensive Gastropathie||||x||x||||||||||||
| |
| − | |-
| |
| − | |Pseudomyxoma peritonei||x||x||||||||||||||
| |
| − | |-
| |
| − | |Pseudozyste||x||||||||||||||||
| |
| − | |-
| |
| − | |Radiaere Narbe||x||x||||||||||||||
| |
| − | |-
| |
| − | |Radioderm||||||x||||||||||||
| |
| − | |-
| |
| − | |Radiographie liegt vor||x||x||||||||||||||
| |
| − | |-
| |
| − | |Referenzbegutachtung||x||||||||||||||||
| |
| − | |-
| |
| − | |Referenzbegutachtung Konkordanz||x||x||||||||||||||
| |
| − | |-
| |
| − | |Residua post abortum||x||x||x||||||||||||
| |
| − | |-
| |
| − | |Residua post partum||x||||x||||||||||||
| |
| − | |-
| |
| − | |Rokitansky-Aschoff-Sinus||x||x||||||||||||||
| |
| − | |-
| |
| − | |Schleimhaut passt zum Zyklustermin||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Schrumpfniere vaskulaer||||||x||||||||||||
| |
| − | |-
| |
| − | |Sehne Degeneration||x||||x||||||||||||
| |
| − | |-
| |
| − | |Sehne Ruptur||x||||x||||||||||||
| |
| − | |-
| |
| − | |Sehne Ruptur klinisch||x||||||||||||||||
| |
| − | |-
| |
| − | |Sertoli cell only Syndrom||x||||||||||||||||
| |
| − | |-
| |
| − | |Sertolizellknoetchen||x||||||||||||||||
| |
| − | |-
| |
| − | |Siderose||x||||||||||||||||
| |
| − | |-
| |
| − | |Solitaere Nabelschnurarterie||||x||||||||||||||
| |
| − | |-
| |
| − | |Trauma||||||x||||||||||||
| |
| − | |-
| |
| − | |Trauma klinisch||x||||x||||||||||||
| |
| − | |-
| |
| − | |Trauma Ruptur||||||x||||||||||||
| |
| − | |-
| |
| − | |Ulcus||x||x||||||||||||||
| |
| − | |-
| |
| − | |Ulcus Erosion||||x||||||||||||||
| |
| − | |-
| |
| − | |Ulcus Erosion inkomplett||x||x||||||||||||||
| |
| − | |-
| |
| − | |Ulcus Erosion komplett||x||x||||||||||||||
| |
| − | |-
| |
| − | |Vollstaendig circulaer||x||x||||x||||||||||
| |
| − | |-
| |
| − | |Zyste||||||x||||||||||||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 20: Attribute mit booleschen Werten (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | Die Informationen können dann gemäß der nachfolgenden Tabelle übermittelt werden:
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Wert!!Datentyp!!Darstellung
| |
| − | |-
| |
| − | |ja||BL||value=\’true\’
| |
| − | |-
| |
| − | |nein||BL||value=\’false\’
| |
| − | |-
| |
| − | |unbekannt||BL||nullFlavor=\’NI\’
| |
| − | |-
| |
| − | |möglich||CD||code=
| |
| − | |-
| |
| − | |unklar||CD||code=
| |
| − | |-
| |
| − | |Verdacht||CD||code=
| |
| − | |-
| |
| − | |nicht untersucht||CD||code=
| |
| − | |-
| |
| − | |nicht beurteilbar||CD||code=
| |
| − | |-
| |
| − | |fraglich|CD||code=
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 21: Darstellung der Attribute mit booleschen Werten (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ==64. Attribut-Code-Paare (noch nicht näher zugeordnet)==
| |
| − | | |
| − | {{AlertBox|Die folgende Tabelle muss auf jeden Fall hinsichtlich der Werte aufgeräumt werden!
| |
| − | Auch ist eine Zuordnung von Datentypen notwendig! In einigen Fällen wird man BL verwenden können. Ansonsten müssen Codesysteme deklariert werden, die z.T. mehrfach genutzt werden können!
| |
| − | Auch werden die Attribute aufgeräumt werden müssen. Fragliche Werte sind in orange markiert.
| |
| − | }}
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Attribut!!Wert
| |
| − | |-
| |
| − | |Abort||Blasenmole möglich
| |
| − | |-
| |
| − | |Abort||Embyonalmole möglich
| |
| − | |-
| |
| − | |Abort||Extrauterin
| |
| − | |-
| |
| − | |Abort||Extrauterin möglich
| |
| − | |-
| |
| − | |Abort||Partialmole möglich
| |
| − | |-
| |
| − | |Abort||Unklar
| |
| − | |-
| |
| − | |Abort||Uterin
| |
| − | |-
| |
| − | |Abort||Uterin möglich
| |
| − | |-
| |
| − | |Abort||Windmole möglich
| |
| − | |-
| |
| − | |Abort klinisch||Blasenmole Verdacht
| |
| − | |-
| |
| − | |Abort klinisch||Embryonalmole Verdacht
| |
| − | |-
| |
| − | |Abort klinisch||Extrauterin
| |
| − | |-
| |
| − | |Abort klinisch||Extrauterin möglich
| |
| − | |-
| |
| − | |Abort klinisch||Partialmole Verdacht
| |
| − | |-
| |
| − | |Abort klinisch||Unbekannt
| |
| − | |-
| |
| − | |Abort klinisch||Uterin
| |
| − | |-
| |
| − | |Abort klinisch||Uterin moeglich
| |
| − | |-
| |
| − | |Abort klinisch||Windmole Verdacht
| |
| − | |-
| |
| − | |Adenose||Blunt duct
| |
| − | |-
| |
| − | |Adenose||Sklerosierend
| |
| − | |-
| |
| − | |Adenose||Sklerosierend nodulaer
| |
| − | |-
| |
| − | |Amputation||negativ
| |
| − | |-
| |
| − | |Amputation||positiv
| |
| − | |-
| |
| − | |Amputation sekundaer||negativ
| |
| − | |-
| |
| − | |Amputation sekundaer||positiv
| |
| − | |-
| |
| − | |Amputation tertiaer||negativ
| |
| − | |-
| |
| − | |Amputation tertiaer||positiv
| |
| − | |-
| |
| − | |Amputation quartaer||negativ
| |
| − | |-
| |
| − | |Amputation quartaer||positiv
| |
| − | |-
| |
| − | |Anastomose||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Anastomose||Insuffizienz Klinisch
| |
| − | |-
| |
| − | |Anastomose||Insuffizienz moeglich
| |
| − | |-
| |
| − | |Anastomose||Regelhaft
| |
| − | |-
| |
| − | |Anastomose||Tumorrezidiv
| |
| − | |-
| |
| − | |Anastomose||Ulcus
| |
| − | |-
| |
| − | |Aneurysma||Ja
| |
| − | |-
| |
| − | |Aneurysma||Klinisch
| |
| − | |-
| |
| − | |Aneurysma||Klinisch Verdacht
| |
| − | |-
| |
| − | |Aneurysma||Moeglich
| |
| − | |-
| |
| − | |Arteriosclerose||Ja
| |
| − | |-
| |
| − | |Arteriosclerose||Klinisch
| |
| − | |-
| |
| − | |Arteriosclerose||Klinisch Verdacht
| |
| − | |-
| |
| − | |Arteriosclerose||Möglich
| |
| − | |-
| |
| − | |Arteriosclerose||Nein
| |
| − | |-
| |
| − | |Arteriovenoese Malformation||Ja
| |
| − | |-
| |
| − | |Arteriovenoese Malformation||Klinisch
| |
| − | |-
| |
| − | |Arteriovenoese Malformation||Moeglich
| |
| − | |-
| |
| − | |Atrophie||Gering
| |
| − | |-
| |
| − | |Atrophie||Gering diffus
| |
| − | |-
| |
| − | |Atrophie||Gering fokal
| |
| − | |-
| |
| − | |Atrophie||Gering ueberwiegend
| |
| − | |-
| |
| − | |Atrophie||Mittelgradig
| |
| − | |-
| |
| − | |Atrophie||Mittelgradig diffus
| |
| − | |-
| |
| − | |Atrophie||Mittelgradig fokal
| |
| − | |-
| |
| − | |Atrophie||Mittelgradig partiell
| |
| − | |-
| |
| − | |Atrophie||Mittelgradig ueberwiegend
| |
| − | |-
| |
| − | |Atrophie||Nein
| |
| − | |-
| |
| − | |Atrophie||Schwergradig
| |
| − | |-
| |
| − | |Atrophie||Schwergradig diffus
| |
| − | |-
| |
| − | |Atrophie||Schwergradig fokal
| |
| − | |-
| |
| − | |Atrophie||Schwergradig partiell
| |
| − | |-
| |
| − | |Atrophie||Schwergradig ueberwiegend
| |
| − | |-
| |
| − | |Auge||Amyloidose
| |
| − | |-
| |
| − | |Ausbreitung||Cervix ja
| |
| − | |-
| |
| − | |Ausbreitung||Cervix nein
| |
| − | |-
| |
| − | |Ausbreitung||Myometrium äußere Hälfte
| |
| − | |-
| |
| − | |Ausbreitung||Myometrium innere Hälfte
| |
| − | |-
| |
| − | |Ausbreitung||Parametrien ja
| |
| − | |-
| |
| − | |Ausbreitung||Parametrien nein
| |
| − | |-
| |
| − | |Barrett-Mucosa||Praebecherzellen
| |
| − | |-
| |
| − | |Beurteilbarkeit||ausreichend
| |
| − | |-
| |
| − | |Beurteilbarkeit||eingeschränkt
| |
| − | |-
| |
| − | |Beurteilbarkeit||gut
| |
| − | |-
| |
| − | |Beurteilbarkeit||unzureichend
| |
| − | |-
| |
| − | |BI-RADS||1
| |
| − | |-
| |
| − | |BI-RADS||2
| |
| − | |-
| |
| − | |BI-RADS||3
| |
| − | |-
| |
| − | |BI-RADS||4
| |
| − | |-
| |
| − | |BI-RADS||4a
| |
| − | |-
| |
| − | |BI-RADS||4b
| |
| − | |-
| |
| − | |BI-RADS||5
| |
| − | |-
| |
| − | |BI-RADS||Unbekannt
| |
| − | |-
| |
| − | |Bowenoide Papulose||Moeglich
| |
| − | |-
| |
| − | |Colon||Divertikel
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis Crohn klinisch
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis Crohn klinisch anamnestisch
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis Crohn klinisch Verdacht
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis Crohn moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis cystica profunda moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis Diversion moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis eosinophil klinisch Verdacht
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis eosinophil moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Bact
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Bact Spirochaetose
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes klinisch
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes klinisch Verdacht
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Parasit
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Parasit Helminth
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Pilz
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Prot Amoebe
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis infektioes Vir CMV
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ischaemisch klinisch
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ischaemisch klinisch Verdacht
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ischaemisch moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis kollagen moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis medikamentoes moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis mikroskopisch moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis pseudomembranoes klinisch
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis pseudomembranoes moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis radiogen moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ulcerosa klinisch
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ulcerosa klinisch Therapie
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ulcerosa klinisch Verdacht
| |
| − | |-
| |
| − | |Colon||Entzuendung Colitis ulcerosa moeglich
| |
| − | |-
| |
| − | |Colon||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Colon||Haemorrhagie
| |
| − | |-
| |
| − | |Colon||Perforation
| |
| − | |-
| |
| − | |Colon||Perforation klinisch
| |
| − | |-
| |
| − | |Colon||Perforation klinisch gedeckt
| |
| − | |-
| |
| − | |Colon||Perforation klinisch Verdacht
| |
| − | |-
| |
| − | |Colon||Perforation moeglich
| |
| − | |-
| |
| − | |Colon||Pseudomelanose
| |
| − | |-
| |
| − | |Colon||Regelhaft
| |
| − | |-
| |
| − | |Colon||Siderose
| |
| − | |-
| |
| − | |Darm Colon Appendix||Appendikopathie neurogen
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis Crohn klinisch
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis Crohn moeglich
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis granulomatoes
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis infektioes moeglich
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis infektioes Parasit
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis infektioes Parasit Helminth
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis ulcerosa klinisch
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung Colitis ulcerosa moeglich
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung unklassifiziert akut
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung unklassifiziert chronisch
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung unklassifiziert chronisch Fibrose
| |
| − | |-
| |
| − | |Darm Colon Appendix||Entzuendung unklassifiziert Rezidiv akut
| |
| − | |-
| |
| − | |Darm Colon Appendix||Perforation
| |
| − | |-
| |
| − | |Darm Colon Appendix||Perforation klinisch
| |
| − | |-
| |
| − | |Darm Colon Appendix||Perforation klinisch Verdacht
| |
| − | |-
| |
| − | |Darm Colon Appendix||Perforation moeglich
| |
| − | |-
| |
| − | |Darm Colon Appendix||Perforation nein
| |
| − | |-
| |
| − | |Darm Duenndarm||Entzuendung Colitis Crohn klinisch anamnestisch
| |
| − | |-
| |
| − | |Darm Duenndarm||Regelhaft
| |
| − | |-
| |
| − | |Darm Duenndarm Duodenum||Amyloidose
| |
| − | |-
| |
| − | |Darm Duenndarm Duodenum||Regelhaft
| |
| − | |-
| |
| − | |Darm Duenndarm Duodenum Papille||Regelhaft
| |
| − | |-
| |
| − | |Differenzierung||Mesonephrogen
| |
| − | |-
| |
| − | |Differenzierung Basalzelle||Ja
| |
| − | |-
| |
| − | |Ductus urachus||Negativ
| |
| − | |-
| |
| − | |Ductus urachus||Nicht untersucht
| |
| − | |-
| |
| − | |Ductus urachus||Positiv
| |
| − | |-
| |
| − | |Duenndarm||Divertikel
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Colitis ulcerosa klinisch
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Colitis ulcerosa klinisch Verdacht
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Crohn klinisch
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Crohn klinisch Verdacht
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Crohn moeglich
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung infektioes Bact Mycobacteriose
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung infektioes klinisch Verdacht
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung infektioes moeglich
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung infektioes Prot Amoebe
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung ischaemisch klinisch
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung ischaemisch klinisch Verdacht
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung ischaemisch moeglich
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung medikamentoes klinisch moeglich
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung medikamentoes moeglich
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Peritonitis
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Peritonitis Fremdkoerper
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Sprue kollagen
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung Sprue kollagen moeglich
| |
| − | |-
| |
| − | |Duenndarm||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Duenndarm||Haemorrhagie
| |
| − | |-
| |
| − | |Duenndarm||Lymphangiektasie ja
| |
| − | |-
| |
| − | |Duenndarm||Lymphangiektasie nein
| |
| − | |-
| |
| − | |Duenndarm||Perforation
| |
| − | |-
| |
| − | |Duenndarm Duodenum||Entzuendung Crohn moeglich
| |
| − | |-
| |
| − | |Durchmesser||unbekannt
| |
| − | |-
| |
| − | |Durchmesser||Unklar
| |
| − | |-
| |
| − | |Durchmesser klinisch||Unbekannt
| |
| − | |-
| |
| − | |Ektomie||negativ
| |
| − | |-
| |
| − | |Ektomie||positiv
| |
| − | |-
| |
| − | |Ektomie CIS||positiv
| |
| − | |-
| |
| − | |Ektopie||Endosalpingiose
| |
| − | |-
| |
| − | |Ektopie||Ja
| |
| − | |-
| |
| − | |Ektopie||Naevus melanozytaer
| |
| − | |-
| |
| − | |Entnahme||Abort
| |
| − | |-
| |
| − | |Entnahme||Abradat
| |
| − | |-
| |
| − | |Entnahme||Abradat klinisch post Abortum
| |
| − | |-
| |
| − | |Entnahme||Abradat klinisch post Partum
| |
| − | |-
| |
| − | |Entnahme||Abstrich
| |
| − | |-
| |
| − | |Entnahme||Amputation
| |
| − | |-
| |
| − | |Entnahme||Amputation quartaer
| |
| − | |-
| |
| − | |Entnahme||Amputation sekundaer
| |
| − | |-
| |
| − | |Entnahme||Amputation tertiaer
| |
| − | |-
| |
| − | |Entnahme||Aquadissektion
| |
| − | |-
| |
| − | |Entnahme||Biopsie
| |
| − | |-
| |
| − | |Entnahme||Biopsie ipsilateral
| |
| − | |-
| |
| − | |Entnahme||Biopsie kontralateral
| |
| − | |-
| |
| − | |Entnahme||Biopsie sekundaer
| |
| − | |-
| |
| − | |Entnahme||Chordektomie
| |
| − | |-
| |
| − | |Entnahme||Colostoma Anus praeter
| |
| − | |-
| |
| − | |Entnahme||Conisch ja
| |
| − | |-
| |
| − | |Entnahme||Conisch nein
| |
| − | |-
| |
| − | |Entnahme||Ektomie
| |
| − | |-
| |
| − | |Entnahme||Ektomie Coblation
| |
| − | |-
| |
| − | |Entnahme||Ektomie laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||Ektomie Residualtumor
| |
| − | |-
| |
| − | |Entnahme||Ektomie sekundaer
| |
| − | |-
| |
| − | |Entnahme||En bloc
| |
| − | |-
| |
| − | |Entnahme||Enukleat
| |
| − | |-
| |
| − | |Entnahme||Enukleat laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||Exprimat
| |
| − | |-
| |
| − | |Entnahme||Exzision
| |
| − | |-
| |
| − | |Entnahme||Exzision transanal
| |
| − | |-
| |
| − | |Entnahme||Fragmentiert
| |
| − | |-
| |
| − | |Entnahme||Hemihepatektomie
| |
| − | |-
| |
| − | |Entnahme||Herniotomie
| |
| − | |-
| |
| − | |Entnahme||Herniotomie laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||Ileostoma
| |
| − | |-
| |
| − | |Entnahme||Ileostoma Anus praeter
| |
| − | |-
| |
| − | |Entnahme||Intakt
| |
| − | |-
| |
| − | |Entnahme||Kapsel
| |
| − | |-
| |
| − | |Entnahme||Konus
| |
| − | |-
| |
| − | |Entnahme||Laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||Lobektomie
| |
| − | |-
| |
| − | |Entnahme||Longo
| |
| − | |-
| |
| − | |Entnahme||Loop excision
| |
| − | |-
| |
| − | |Entnahme||Lymphknoten primaer
| |
| − | |-
| |
| − | |Entnahme||Lymphknoten sekundaer
| |
| − | |-
| |
| − | |Entnahme||Mastektomie
| |
| − | |-
| |
| − | |Entnahme||Mastektomie sekundaer
| |
| − | |-
| |
| − | |Entnahme||Mastektomie tertiaer
| |
| − | |-
| |
| − | |Entnahme||Morcellement
| |
| − | |-
| |
| − | |Entnahme||Mukosektomie
| |
| − | |-
| |
| − | |Entnahme||Neck dissection
| |
| − | |-
| |
| − | |Entnahme||Phlebotomie
| |
| − | |-
| |
| − | |Entnahme||PME laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||PME partielle mesorectale Excision
| |
| − | |-
| |
| − | |Entnahme||Pneumektomie
| |
| − | |-
| |
| − | |Entnahme||Polyp
| |
| − | |-
| |
| − | |Entnahme||Punktat
| |
| − | |-
| |
| − | |Entnahme||Reduktionsplastik
| |
| − | |-
| |
| − | |Entnahme||Resektat
| |
| − | |-
| |
| − | |Entnahme||Resektat laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||Resektat multipel
| |
| − | |-
| |
| − | |Entnahme||Resektat primaer
| |
| − | |-
| |
| − | |Entnahme||Resektat quartaer
| |
| − | |-
| |
| − | |Entnahme||Resektat quintaer
| |
| − | |-
| |
| − | |Entnahme||Resektat sekundaer
| |
| − | |-
| |
| − | |Entnahme||Resektat subtotal
| |
| − | |-
| |
| − | |Entnahme||Resektat tertiaer
| |
| − | |-
| |
| − | |Entnahme||Resektat Vollwand
| |
| − | |-
| |
| − | |Entnahme||Resektat Whipple
| |
| − | |-
| |
| − | |Entnahme||Sectio
| |
| − | |-
| |
| − | |Entnahme||Spontan ausgestossen
| |
| − | |-
| |
| − | |Entnahme||Stanze
| |
| − | |-
| |
| − | |Entnahme||Stanze Mammotom
| |
| − | |-
| |
| − | |Entnahme||Stanze Vakuum
| |
| − | |-
| |
| − | |Entnahme||Thrombendarteriektomie
| |
| − | |-
| |
| − | |Entnahme||TME laparoskopisch
| |
| − | |-
| |
| − | |Entnahme||TME mit Amputation
| |
| − | |-
| |
| − | |Entnahme||TME vollstaendige mesorectale Excision
| |
| − | |-
| |
| − | |Entnahme||Tumorektomie TMMR
| |
| − | |-
| |
| − | |Entnahme||Unbekannt
| |
| − | |-
| |
| − | |Entnahme||Unvollstaendig
| |
| − | |-
| |
| − | |Entnahme||Zerklueftet
| |
| − | |-
| |
| − | |Entnahme Lymphknoten||Ektomie
| |
| − | |-
| |
| − | |Entnahme Lymphknoten||Ektomie laparoskopisch
| |
| − | |-
| |
| − | |Entzuendung||Abscess
| |
| − | |-
| |
| − | |Entzuendung||Abszess
| |
| − | |-
| |
| − | |Entzuendung||Abszess Akne inversa moeglich
| |
| − | |-
| |
| − | |Entzuendung||Abszess Sinus pilonidalis
| |
| − | |-
| |
| − | |Entzuendung||Acut
| |
| − | |-
| |
| − | |Entzuendung||Akut
| |
| − | |-
| |
| − | |Entzuendung||Akut Abscess
| |
| − | |-
| |
| − | |Entzuendung||Akut Abszess
| |
| − | |-
| |
| − | |Entzuendung||Akut Recidiv
| |
| − | |-
| |
| − | |Entzuendung||Akut Rezidiv unklassifiziert
| |
| − | |-
| |
| − | |Entzuendung||Akut unklassifiziert
| |
| − | |-
| |
| − | |Entzuendung||Akut Unklassifiziert Minimalbild
| |
| − | |-
| |
| − | |Entzuendung||Akut Unklassifiziert Teilbild
| |
| − | |-
| |
| − | |Entzuendung||Akut Unklassifiziert Vollbild
| |
| − | |-
| |
| − | |Entzuendung||Aortenaneurysma inflammatoridsch moeglich
| |
| − | |-
| |
| − | |Entzuendung||Bact Aktinomykose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Bact Spirochaetose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Bact Whipple moeglich
| |
| − | |-
| |
| − | |Entzuendung||Bakerzyste moeglich
| |
| − | |-
| |
| − | |Entzuendung||Bursitis akut
| |
| − | |-
| |
| − | |Entzuendung||Bursitis chronisch
| |
| − | |-
| |
| − | |Entzuendung||Chorionamnionitis
| |
| − | |-
| |
| − | |Entzuendung||Chronisch
| |
| − | |-
| |
| − | |Entzuendung||Chronisch Eosinophilie
| |
| − | |-
| |
| − | |Entzuendung||Chronisch Fibrose
| |
| − | |-
| |
| − | |Entzuendung||Chronisch Narbe
| |
| − | |-
| |
| − | |Entzuendung||Chronisch Recidiv acut
| |
| − | |-
| |
| − | |Entzuendung||Chronisch sclerosierend
| |
| − | |-
| |
| − | |Entzuendung||Chronisch Ulcus
| |
| − | |-
| |
| − | |Entzuendung||Chronisch unklassifiziert
| |
| − | |-
| |
| − | |Entzuendung||Desmet
| |
| − | |-
| |
| − | |Entzuendung||Drusen
| |
| − | |-
| |
| − | |Entzuendung||Eitrig Abszess
| |
| − | |-
| |
| − | |Entzuendung||Eitrig Fistel
| |
| − | |-
| |
| − | |Entzuendung||Ekzem dyshidrosiform moeglich
| |
| − | |-
| |
| − | |Entzuendung||Ekzem moeglich
| |
| − | |-
| |
| − | |Entzuendung||Entzuendung Aktinomykose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Entzuendung Sarkoidose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Entzuendung Tuberkulose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Eosinophil
| |
| − | |-
| |
| − | |Entzuendung||Erysipel moeglich
| |
| − | |-
| |
| − | |Entzuendung||Erythema nodosum moeglich
| |
| − | |-
| |
| − | |Entzuendung||Fasziitis nekrotisierend moeglich
| |
| − | |-
| |
| − | |Entzuendung||Fibromatose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Fibromatose retroperitoneal moeglilch
| |
| − | |-
| |
| − | |Entzuendung||Fistel
| |
| − | |-
| |
| − | |Entzuendung||Fistel Akne inversa moeglich
| |
| − | |-
| |
| − | |Entzuendung||Fistel Crohn moeglich
| |
| − | |-
| |
| − | |Entzuendung||Fistel Sinus pilonidalis
| |
| − | |-
| |
| − | |Entzuendung||Fremdkoerper
| |
| − | |-
| |
| − | |Entzuendung||Fremdkoerper Kunststoff
| |
| − | |-
| |
| − | |Entzuendung||Fremdkoerper Paraffin moeglich
| |
| − | |-
| |
| − | |Entzuendung||Funtkionssteigerung
| |
| − | |-
| |
| − | |Entzuendung||Gangraen Fournier moeglich
| |
| − | |-
| |
| − | |Entzuendung||Graft versus host moeglich
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes aktinisch elastolytisch
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes BCG Verdacht
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Sarkoidosetyp
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Sarkoidose-Typ
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Spermagranulom
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Tuberkulose
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Tuberkulose klinisch Therapie
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Tuberkulose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Tuberkulose Verdacht
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Tuberkulosetyp
| |
| − | |-
| |
| − | |Entzuendung||Granulomatoes Tuberkulose-Typ
| |
| − | |-
| |
| − | |Entzuendung||Hydronephrose
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Bact
| |
| − | |-
| |
| − | |Entzuendung||Infektioes moeglich
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Parasit Bilharziose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Parasit Scabies moeglich
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Pilz
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Vir
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Vir EBV moeglich
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Vir Herpes
| |
| − | |-
| |
| − | |Entzuendung||Infektioes Vir moeglich
| |
| − | |-
| |
| − | |Entzuendung||Kollagenose perforierend moeglich
| |
| − | |-
| |
| − | |Entzuendung||Lichen ruber klinisch Verdacht
| |
| − | |-
| |
| − | |Entzuendung||Lichen ruber moeglich
| |
| − | |-
| |
| − | |Entzuendung||Lichen sclerosus et atrophicus
| |
| − | |-
| |
| − | |Entzuendung||Lichenoide Keratose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Lupus erythematodes moeglich
| |
| − | |-
| |
| − | |Entzuendung||Lymphadenopathie
| |
| − | |-
| |
| − | |Entzuendung||Lymphadenopathie dermatopathisch
| |
| − | |-
| |
| − | |Entzuendung||Lymphadenopathie Epitheloidzellreaktion
| |
| − | |-
| |
| − | |Entzuendung||Lymphadenopathie Toxoplasmose moeglich
| |
| − | |-
| |
| − | |Entzuendung||Nekrose
| |
| − | |-
| |
| − | |Entzuendung||Nekrose fibrinoid
| |
| − | |-
| |
| − | |Entzuendung||Pankreatitis segmental
| |
| − | |-
| |
| − | |Entzuendung||Panniculitis
| |
| − | |-
| |
| − | |Entzuendung||Perforation moeglich
| |
| − | |-
| |
| − | |Entzuendung||Perinephritis akut Fistel
| |
| − | |-
| |
| − | |Entzuendung||Perinephritis unklassifiziert
| |
| − | |-
| |
| − | |Entzuendung||Periorchitis nodulaer moeglich
| |
| − | |-
| |
| − | |Entzuendung||Peritonitis
| |
| − | |-
| |
| − | |Entzuendung||Peritonitis Fremdkoerper
| |
| − | |-
| |
| − | |Entzuendung||Peritonitis Pilz
| |
| − | |-
| |
| − | |Entzuendung||Phlegmone
| |
| − | |-
| |
| − | |Entzuendung||Pilz
| |
| − | |-
| |
| − | |Entzuendung||Plasmacellularis Zoon
| |
| − | |-
| |
| − | |Entzuendung||Plasmacellularis Zoon Klinisch Vedacht
| |
| − | |-
| |
| − | |Entzuendung||Plasmacellularis Zoon moeglich
| |
| − | |-
| |
| − | |Entzuendung||Prot Cryptosporidien moeglich
| |
| − | |-
| |
| − | |Entzuendung||Prot Lamblien
| |
| − | |-
| |
| − | |Entzuendung||Prot Lamblien moeglich
| |
| − | |-
| |
| − | |Entzuendung||Prurigo simplex moeglich
| |
| − | |-
| |
| − | |Entzuendung||Pseudotumor fibroes moeglich
| |
| − | |-
| |
| − | |Entzuendung||Pseudotumor inflammatorisch moeglich
| |
| − | |-
| |
| − | |Entzuendung||Psoriasiform
| |
| − | |-
| |
| − | |Entzuendung||Psoriasis moeglich
| |
| − | |-
| |
| − | |Entzuendung||Radiogen moeglich
| |
| − | |-
| |
| − | |Entzuendung||Recidiv akut
| |
| − | |-
| |
| − | |Entzuendung||Rinnenpankreatitis
| |
| − | |-
| |
| − | |Entzuendung||Serom moeglich
| |
| − | |-
| |
| − | |Entzuendung||Ulcus
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiert
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert akut
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert akut Abszess
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert akut Peritonsillarabszess
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert akut Rezidiv Ulcus
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert chronisch
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert chronisch Fibrose
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert chronisch floride
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert chronisch Rezidiv akut
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert floride
| |
| − | |-
| |
| − | |Entzuendung||Unklassifiziert Rezidiv akut
| |
| − | |-
| |
| − | |Entzuendung||Unklassifziert
| |
| − | |-
| |
| − | |Entzuendung||Unklassifziert akut
| |
| − | |-
| |
| − | |Entzuendung||Unklassifziert akut Abszess
| |
| − | |-
| |
| − | |Entzuendung||Unklassifziert chronisch
| |
| − | |-
| |
| − | |Entzuendung||Urtikaria moeglich
| |
| − | |-
| |
| − | |Entzuendung||Vaskulitis
| |
| − | |-
| |
| − | |Entzuendung||Vaskulitis leukozytoklastisch moeglich
| |
| − | |-
| |
| − | |Entzuendung||Vaskulitis moeglich
| |
| − | |-
| |
| − | |Entzuendung||Wundreaktion
| |
| − | |-
| |
| − | |Entzuendung||Wundreaktion Spindelzellproliferation
| |
| − | |-
| |
| − | |Entzuendung klinisch||Abscess
| |
| − | |-
| |
| − | |Entzuendung klinisch||Erysipel
| |
| − | |-
| |
| − | |Entzuendung klinisch||Fasziitis nekrotisierend Verdacht
| |
| − | |-
| |
| − | |Entzuendung klinisch||Fibromatose retroperitoneal Verdacht
| |
| − | |-
| |
| − | |Entzuendung klinisch||Hydronephrose
| |
| − | |-
| |
| − | |Entzuendung klinisch||Lupus erythematodes Verdacht
| |
| − | |-
| |
| − | |Entzuendung klinisch||Radiogen Verdacht
| |
| − | |-
| |
| − | |Entzuendung klinisch||Sepsis
| |
| − | |-
| |
| − | |Entzuendung klinisch||Vaskulitis Verdacht
| |
| − | |-
| |
| − | |Entzuendung klinisch||Aortenaneurysma inflammatoridsch Verdacht
| |
| − | |-
| |
| − | |Erythrozyten||gering
| |
| − | |-
| |
| − | |Erythrozyten||mittelgradig
| |
| − | |-
| |
| − | |EWGBSP04||B1a
| |
| − | |-
| |
| − | |EWGBSP04||B1b
| |
| − | |-
| |
| − | |EWGBSP04||B2
| |
| − | |-
| |
| − | |EWGBSP04||B3
| |
| − | |-
| |
| − | |EWGBSP04||B4
| |
| − | |-
| |
| − | |EWGBSP04||B5
| |
| − | |-
| |
| − | |EWGBSP04||B5a
| |
| − | |-
| |
| − | |EWGBSP04||B5b
| |
| − | |-
| |
| − | |EWGBSP04||B5c
| |
| − | |-
| |
| − | |EWGBSP04||B5d
| |
| − | |-
| |
| − | |EWGBSP04||C2
| |
| − | |-
| |
| − | |EWGBSP04||C3
| |
| − | |-
| |
| − | |EWGBSP04||C4
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B1a
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B1b
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B2
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B3
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B4
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B5a
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B5b
| |
| − | |-
| |
| − | |EWGBSP04 Referenzpathologie||B5c
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B1a
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B1b
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B2
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B3
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B4
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B5
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B5a
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B5b
| |
| − | |-
| |
| − | |EWGBSP04-Schnellschnitt||B5c
| |
| − | |-
| |
| − | |Fascie||Eingeschnitten
| |
| − | |-
| |
| − | |Fascie||Intact
| |
| − | |-
| |
| − | |Fascie||Mesorectum zerklueftet
| |
| − | |-
| |
| − | |Fascie||Ruptur gross
| |
| − | |-
| |
| − | |Fascie||Ruptur gross Muscularis
| |
| − | |-
| |
| − | |Fascie||Ruptur klein
| |
| − | |-
| |
| − | |Faszie basal||Ja
| |
| − | |-
| |
| − | |Faszie basal||Nein
| |
| − | |-
| |
| − | |Faszie basal||Unklar
| |
| − | |-
| |
| − | |Faszie basal markiert||Ja
| |
| − | |-
| |
| − | |Faszie basal markiert||Nein
| |
| − | |-
| |
| − | |Fehlbildung||Moeglich
| |
| − | |-
| |
| − | |Fehlbildung klinisch||Verdacht
| |
| − | |-
| |
| − | |Fibrose||Desmet
| |
| − | |-
| |
| − | |Fibroxanthom atypisch||Ja
| |
| − | |-
| |
| − | |Fraktur||Kallus bindegewebig
| |
| − | |-
| |
| − | |Fraktur||Kallus knoechern
| |
| − | |-
| |
| − | |Fraktur||Moeglich
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Cholesteatose
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Cholesteatose polypös
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Lithiasis ja
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Lithiasis klinisch ja
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Lithiasis klinisch nein
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Lithiasis klinisch unbekannt
| |
| − | |-
| |
| − | |Gallenweg Gallenblase||Lithiasis nein
| |
| − | |-
| |
| − | |Gastritis||A
| |
| − | |-
| |
| − | |Gastritis||A/B
| |
| − | |-
| |
| − | |Gastritis||Anastomose
| |
| − | |-
| |
| − | |Gastritis||Anastomose B-1
| |
| − | |-
| |
| − | |Gastritis||Anastomose B-2
| |
| − | |-
| |
| − | |Gastritis||B
| |
| − | |-
| |
| − | |Gastritis||C
| |
| − | |-
| |
| − | |Gastritis||Crohn moeglich
| |
| − | |-
| |
| − | |Gastritis||granulomatoes
| |
| − | |-
| |
| − | |Gastritis||nein
| |
| − | |-
| |
| − | |Gastritis||unklassifiziert
| |
| − | |-
| |
| − | |Gefaess||Entzuendung riesenzellig Horton klinisch
| |
| − | |-
| |
| − | |Gefaess||Entzuendung riesenzellig Horton klinisch Verdacht
| |
| − | |-
| |
| − | |Gefaess||Entzuendung riesenzellig Horton moeglich
| |
| − | |-
| |
| − | |Gefaess||Entzuendung Vasculitis moeglich
| |
| − | |-
| |
| − | |Gefaessausbreitung Lokalisation||Extratumoral
| |
| − | |-
| |
| − | |Gefaessausbreitung Lokalisation||Intratumoral
| |
| − | |-
| |
| − | |Gelenk||Arthropathie Amyloid
| |
| − | |-
| |
| − | |Gelenk||Arthropathie Kalk
| |
| − | |-
| |
| − | |Gelenk||Arthropathie Kristalle
| |
| − | |-
| |
| − | |Gelenk||Arthropathie Kristalle Pyrophosphat moeglich
| |
| − | |-
| |
| − | |Gelenk||Arthropathie Kristalle Urat moeglich
| |
| − | |-
| |
| − | |Gelenk||Entzuendung infektioes Bact
| |
| − | |-
| |
| − | |Gelenk||Entzuendung Siderose
| |
| − | |-
| |
| − | |Gelenk||Entzündung eitrig
| |
| − | |-
| |
| − | |Gelenk||Entzündung Prothese
| |
| − | |-
| |
| − | |Gelenk||Entzündung rheumoatoid moeglich
| |
| − | |-
| |
| − | |Gelenk||Entzündung Tendovaginitis de Quervain moeglich
| |
| − | |-
| |
| − | |Gelenk||Entzündung unklassifiziert
| |
| − | |-
| |
| − | |Gelenk||Klinisch Gicht
| |
| − | |-
| |
| − | |Gewebe reif||Ueberwiegend
| |
| − | |-
| |
| − | |Gewicht||Unbekannt
| |
| − | |-
| |
| − | |Gleason||7a
| |
| − | |-
| |
| − | |Gleason||7b
| |
| − | |-
| |
| − | |Haemorrhagie||Aelter
| |
| − | |-
| |
| − | |Haemorrhagie||Eisen ja
| |
| − | |-
| |
| − | |Haemorrhagie||Eisen nein
| |
| − | |-
| |
| − | |Haemorrhagie||Frisch
| |
| − | |-
| |
| − | |Haemorrhagie||Frischer
| |
| − | |-
| |
| − | |Haemorrhagie||Ja
| |
| − | |-
| |
| − | |Haemorrhagie||Organisat unvollstaendig
| |
| − | |-
| |
| − | |Haemorrhagie aelter||Ja
| |
| − | |-
| |
| − | |Haemorrhagie klinisch||Subdural chronisch
| |
| − | |-
| |
| − | |Harnblase||Entzuendung Cystitis interstitiell moeglich
| |
| − | |-
| |
| − | |Harnblase||Entzuendung granulomatoes
| |
| − | |-
| |
| − | |Harnblase||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Harnblase||Intact
| |
| − | |-
| |
| − | |Harnblase||Regelhaft
| |
| − | |-
| |
| − | |Harnblase||Ruptur
| |
| − | |-
| |
| − | |Haut||Gicht
| |
| − | |-
| |
| − | |Haut||Regelhaft
| |
| − | |-
| |
| − | |Haut||Siderose
| |
| − | |-
| |
| − | |HELLP-Syndrom||Ja
| |
| − | |-
| |
| − | |HELLP-Syndrom||Moeglich
| |
| − | |-
| |
| − | |Heterotopie||Gastral
| |
| − | |-
| |
| − | |Heterotopie||Pankreatisch
| |
| − | |-
| |
| − | |Hiluszelle||Hyperplasie
| |
| − | |-
| |
| − | |Histologie Marsh||0
| |
| − | |-
| |
| − | |Histologie Marsh||1
| |
| − | |-
| |
| − | |Histologie Marsh||3a
| |
| − | |-
| |
| − | |Histologie Marsh||3b
| |
| − | |-
| |
| − | |Histologie Marsh||3c
| |
| − | |-
| |
| − | |Histologie Marsh||Unklar
| |
| − | |-
| |
| − | |Hyperplasie||adenomatoes atypisch
| |
| − | |-
| |
| − | |Hyperplasie||Glandulaer
| |
| − | |-
| |
| − | |Hyperplasie||Glandulaer Stroma
| |
| − | |-
| |
| − | |Hyperplasie||Stroma
| |
| − | |-
| |
| − | |Involution||Ja
| |
| − | |-
| |
| − | |Involution||Lipomatoes
| |
| − | |-
| |
| − | |Keimepithel||Desquamiert
| |
| − | |-
| |
| − | |Keimepithel||Desquamiert partiell
| |
| − | |-
| |
| − | |Keimepithel||Intakt
| |
| − | |-
| |
| − | |Klinik||Barre
| |
| − | |-
| |
| − | |Klinik Lokalisation||Mammographie
| |
| − | |-
| |
| − | |Klinik Lokalisation||MRT
| |
| − | |-
| |
| − | |Klinik Lokalisation||Palpation
| |
| − | |-
| |
| − | |Klinik Lokalisation||Sonographie
| |
| − | |-
| |
| − | |Klinik Lokalisation||Stereotaxie
| |
| − | |-
| |
| − | |Klinik Lokalisation||Unbekannt
| |
| − | |-
| |
| − | |Klinik Marsh||0
| |
| − | |-
| |
| − | |Klinik Marsh||1
| |
| − | |-
| |
| − | |Klinik Marsh||2
| |
| − | |-
| |
| − | |Klinik Marsh||3
| |
| − | |-
| |
| − | |Klinik Marsh||3a
| |
| − | |-
| |
| − | |Klinik Marsh||3c
| |
| − | |-
| |
| − | |Klinik Marsh||unbekannt
| |
| − | |-
| |
| − | |Klinik PET||Negativ
| |
| − | |-
| |
| − | |Klinik PET||Positiv
| |
| − | |-
| |
| − | |Klinik Zyklustermin||Postmenopausenblutung
| |
| − | |-
| |
| − | |Klinik Zyklustermin||Unbekannt
| |
| − | |-
| |
| − | |Klinisch||Abszess
| |
| − | |-
| |
| − | |Klinisch||Actinomycose Verdacht
| |
| − | |-
| |
| − | |Klinisch||Anaemie
| |
| − | |-
| |
| − | |Klinisch||Appendix Torsion Verdacht
| |
| − | |-
| |
| − | |Klinisch||Azoospermie
| |
| − | |-
| |
| − | |Klinisch||Bakerzyste
| |
| − | |-
| |
| − | |Klinisch||Bakerzyste Recidiv
| |
| − | |-
| |
| − | |Klinisch||Bakerzyste Verdacht
| |
| − | |-
| |
| − | |Klinisch||Bilharziose Verdacht
| |
| − | |-
| |
| − | |Klinisch||Blutung ja
| |
| − | |-
| |
| − | |Klinisch||Blutung unbekannt
| |
| − | |-
| |
| − | |Klinisch||Bursitis akut
| |
| − | |-
| |
| − | |Klinisch||Bursitis chronisch
| |
| − | |-
| |
| − | |Klinisch||Diarrhoe
| |
| − | |-
| |
| − | |Klinisch||Diarrhoediagnostik
| |
| − | |-
| |
| − | |Klinisch||Divertikel ja
| |
| − | |-
| |
| − | |Klinisch||Divertikel nein
| |
| − | |-
| |
| − | |Klinisch||Divertikel unbekannt
| |
| − | |-
| |
| − | |Klinisch||Entzuendung Cystitis interstitiell unbekannt
| |
| − | |-
| |
| − | |Klinisch||Entzuendungsdiagnostik
| |
| − | |-
| |
| − | |Klinisch||Fertilitaetsdiagnostik
| |
| − | |-
| |
| − | |Klinisch||Fissur
| |
| − | |-
| |
| − | |Klinisch||Fistel
| |
| − | |-
| |
| − | |Klinisch||Funiculocele Verdacht
| |
| − | |-
| |
| − | |Klinisch||Gerinnungsstoerung
| |
| − | |-
| |
| − | |Klinisch||Hydrocele Verdacht
| |
| − | |-
| |
| − | |Klinisch||Hydrops
| |
| − | |-
| |
| − | |Klinisch||Hypoplasie
| |
| − | |-
| |
| − | |Klinisch||Ileus
| |
| − | |-
| |
| − | |Klinisch||Infect Vir Verdacht
| |
| − | |-
| |
| − | |Klinisch||Infektioes Pilz Verdacht
| |
| − | |-
| |
| − | |Klinisch||Kontrolle
| |
| − | |-
| |
| − | |Klinisch||Megaureter
| |
| − | |-
| |
| − | |Klinisch||MRSA
| |
| − | |-
| |
| − | |Klinisch||Multiple Sklerose
| |
| − | |-
| |
| − | |Klinisch||Nephrostomie
| |
| − | |-
| |
| − | |Klinisch||Nierendegeneration polycystisch
| |
| − | |-
| |
| − | |Klinisch||Perforation
| |
| − | |-
| |
| − | |Klinisch||Plasmacellularis Zoon Verdacht
| |
| − | |-
| |
| − | |Klinisch||Pneumothorax
| |
| − | |-
| |
| − | |Klinisch||Primaertumor unbekannt
| |
| − | |-
| |
| − | |Klinisch||Prolaps
| |
| − | |-
| |
| − | |Klinisch||Sarkoidose Verdacht
| |
| − | |-
| |
| − | |Klinisch||Schrumpfharnblase
| |
| − | |-
| |
| − | |Klinisch||Schrumpfniere
| |
| − | |-
| |
| − | |Klinisch||Schrumpfniere Pyelonephritis
| |
| − | |-
| |
| − | |Klinisch||Schrumpfniere vaskulaer
| |
| − | |-
| |
| − | |Klinisch||Schrumpfniere vaskulaer Arterienstenose
| |
| − | |-
| |
| − | |Klinisch||Sinus pilonidalis Rezidiv
| |
| − | |-
| |
| − | |Klinisch||Spermatocele Verdacht
| |
| − | |-
| |
| − | |Klinisch||Sprue
| |
| − | |-
| |
| − | |Klinisch||Sprue Auschlussdiagnostik
| |
| − | |-
| |
| − | |Klinisch||Sprue Verdacht
| |
| − | |-
| |
| − | |Klinisch||Stenose
| |
| − | |-
| |
| − | |Klinisch||Trauma
| |
| − | |-
| |
| − | |Klinisch||Trauma iatrogen
| |
| − | |-
| |
| − | |Klinisch||Trauma unbekannt
| |
| − | |-
| |
| − | |Klinisch||Tuberkulose Verdacht
| |
| − | |-
| |
| − | |Klinisch||Tumordiagnostik
| |
| − | |-
| |
| − | |Klinisch||Tumorverdacht
| |
| − | |-
| |
| − | |Klinisch||Ulcus
| |
| − | |-
| |
| − | |Klinisch||Ureterabgangsstenose
| |
| − | |-
| |
| − | |Klinisch||Vorsorge
| |
| − | |-
| |
| − | |Klinisch Gastrale antrale vaskulaere Ektasie||Verdacht
| |
| − | |-
| |
| − | |Klinische Angaben||ausreichend
| |
| − | |-
| |
| − | |Klinische Angaben||keine
| |
| − | |-
| |
| − | |Klinische Angaben||unzureichend
| |
| − | |-
| |
| − | |Klinische Angaben ausreichend||Ja
| |
| − | |-
| |
| − | |Klinische Angaben ausreichend||Nein
| |
| − | |-
| |
| − | |Klinische Kompartementsyndrom||Ja
| |
| − | |-
| |
| − | |Klinische Zyste||Verdacht
| |
| − | |-
| |
| − | |Leukozyten||gering
| |
| − | |-
| |
| − | |Leukozyten||mittelgradig
| |
| − | |-
| |
| − | |Leukozyten||schwergradig
| |
| − | |-
| |
| − | |Leydigzelle Atrophie||Ja
| |
| − | |-
| |
| − | |Leydigzelle Hyperplasie||Ja
| |
| − | |-
| |
| − | |Leydigzelle Hyperplasie nodulaer||Ja
| |
| − | |-
| |
| − | |Leydigzelle regelhaft||Ja
| |
| − | |-
| |
| − | |Lunge||Entzuendung bronchozentrisch
| |
| − | |-
| |
| − | |Lunge||Entzuendung granulomatoes angiozentrisch
| |
| − | |-
| |
| − | |Lunge||Entzuendung granulomatoes Sarkoidose-Typ
| |
| − | |-
| |
| − | |Lunge||Entzuendung granulomatoes Tuberkulose-Typ
| |
| − | |-
| |
| − | |Lunge||Entzuendung infektioes klinisch
| |
| − | |-
| |
| − | |Lunge||Entzuendung infektioes moeglich
| |
| − | |-
| |
| − | |Lunge||Entzuendung infektioes Pilz
| |
| − | |-
| |
| − | |Lunge||Entzuendung infektioes Vir CMV
| |
| − | |-
| |
| − | |Lunge||Entzuendung medikamentoes moeglich
| |
| − | |-
| |
| − | |Lunge||Entzuendung Pleuritis
| |
| − | |-
| |
| − | |Lunge||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Lunge||Infarkt moeglich
| |
| − | |-
| |
| − | |Lunge||Kreislaufstoerung Embolie alt
| |
| − | |-
| |
| − | |Lunge||Kreislaufstoerung Embolie alt moeglich
| |
| − | |-
| |
| − | |Lunge||Kreislaufstoerung moeglich
| |
| − | |-
| |
| − | |Lunge||Kreislaufstoerung Thrombus aelter
| |
| − | |-
| |
| − | |Lunge||Kreislaufstoerung Thrombus alt
| |
| − | |-
| |
| − | |Lunge||Kreislaufstoerung Thrombus frisch
| |
| − | |-
| |
| − | |Lymphknoten Schnellschnitt||negativ
| |
| − | |-
| |
| − | |Lymphknoten Schnellschnitt||positiv
| |
| − | |-
| |
| − | |Lymphknoten Schnellschnitt||positiv Verdacht
| |
| − | |-
| |
| − | |Lymphknoten Schnellschnitt||unklar
| |
| − | |-
| |
| − | |Magen||Haemorrhagie
| |
| − | |-
| |
| − | |Magen||Perforation
| |
| − | |-
| |
| − | |Magen||Regelhaft
| |
| − | |-
| |
| − | |Magen||Siderose
| |
| − | |-
| |
| − | |Makroorchie||Ja
| |
| − | |-
| |
| − | |Masse||Formalin aufgespannt ja
| |
| − | |-
| |
| − | |Masse||Formalin aufgespannt nein
| |
| − | |-
| |
| − | |Masse||Frisch
| |
| − | |-
| |
| − | |Mastektomie||negativ
| |
| − | |-
| |
| − | |Mastektomie||positiv
| |
| − | |-
| |
| − | |Mastektomie sekundaer||negativ
| |
| − | |-
| |
| − | |Mastektomie sekundaer||positiv
| |
| − | |-
| |
| − | |Mastektomie tertiaer||negativ
| |
| − | |-
| |
| − | |Mastektomie tertiaer||positiv
| |
| − | |-
| |
| − | |Mastozytose||Moeglich
| |
| − | |-
| |
| − | |Medikamente||Chemotherapie
| |
| − | |-
| |
| − | |Medikamente||Ja
| |
| − | |-
| |
| − | |Medikamente||NSAR
| |
| − | |-
| |
| − | |Medikamente||NSAR ASS
| |
| − | |-
| |
| − | |Medikamente||Unbekannt
| |
| − | |-
| |
| − | |Medikamente Steroide||Ja
| |
| − | |-
| |
| − | |Medikamente Steroide||Unbekannt
| |
| − | |-
| |
| − | |Melanozyt||Hyperplasie solar
| |
| − | |-
| |
| − | |Metaplasie||Azinaer
| |
| − | |-
| |
| − | |Metaplasie||Glandulaer
| |
| − | |-
| |
| − | |Metaplasie||Intestinal
| |
| − | |-
| |
| − | |Metaplasie||Intestinal Cardia
| |
| − | |-
| |
| − | |Metaplasie||Nephrogen
| |
| − | |-
| |
| − | |Metaplasie||Ossification
| |
| − | |-
| |
| − | |Metaplasie Coelom Keimstrang||Moeglich
| |
| − | |-
| |
| − | |Metaplasie synovial||Ja
| |
| − | |-
| |
| − | |Metaplasie synovial||Nein
| |
| − | |-
| |
| − | |Metaplasiezellen||Ja
| |
| − | |-
| |
| − | |Metaplasiezellen||Nein
| |
| − | |-
| |
| − | |Metastase kapselueberschreitendes Wachstum||Ja
| |
| − | |-
| |
| − | |Metastase kapselueberschreitendes Wachstum||Nein
| |
| − | |-
| |
| − | |Milz||Intakt
| |
| − | |-
| |
| − | |Milz||Regelhaft
| |
| − | |-
| |
| − | |Milz||Ruptur
| |
| − | |-
| |
| − | |Mischflora||Ja
| |
| − | |-
| |
| − | |Mischflora||Nein
| |
| − | |-
| |
| − | |MRT||T2
| |
| − | |-
| |
| − | |MRT||T3
| |
| − | |-
| |
| − | |MRT||T3b
| |
| − | |-
| |
| − | |MRT||T3c
| |
| − | |-
| |
| − | |MRT||unbekannt
| |
| − | |-
| |
| − | |MRT||unklar
| |
| − | |-
| |
| − | |Nachresektat||negativ
| |
| − | |-
| |
| − | |Nachresektat||positiv
| |
| − | |-
| |
| − | |Nachresektat||unklar
| |
| − | |-
| |
| − | |Nachresektat sekundaer||negativ
| |
| − | |-
| |
| − | |Nachresektat sekundaer||positiv
| |
| − | |-
| |
| − | |Nachresektat sekundaer||unklar
| |
| − | |-
| |
| − | |Nachresektat tertiaer||negativ
| |
| − | |-
| |
| − | |Nachresektat tertiaer||positiv
| |
| − | |-
| |
| − | |Naht||Insuffizienz Klinisch
| |
| − | |-
| |
| − | |Naht||Insuffizienz moeglich
| |
| − | |-
| |
| − | |Narbe radiaer||Ja
| |
| − | |-
| |
| − | |Narbe radiaer||Moeglich
| |
| − | |-
| |
| − | |Navikularzellen||Nein
| |
| − | |-
| |
| − | |Nekrose||Aelter Organisation
| |
| − | |-
| |
| − | |Nekrose||Embolie
| |
| − | |-
| |
| − | |Nekrose||Embolie moeglich
| |
| − | |-
| |
| − | |Nekrose||Frisch Granulozytaer demarkiert
| |
| − | |-
| |
| − | |Nekrose||Frisch Haemorrhagie
| |
| − | |-
| |
| − | |Nekrose||Haemorrhagisch
| |
| − | |-
| |
| − | |Nekrose||Haemorrhagische Infarcierung
| |
| − | |-
| |
| − | |Nekrose||Ja
| |
| − | |-
| |
| − | |Nekrose||Klinisch Ileus
| |
| − | |-
| |
| − | |Nekrose||Klinisch Nahtinsuffzienz
| |
| − | |-
| |
| − | |Nekrose||Manifest ja
| |
| − | |-
| |
| − | |Nekrose||Manifest nein
| |
| − | |-
| |
| − | |Nekrose||Moeglich
| |
| − | |-
| |
| − | |Nekrose||Nein
| |
| − | |-
| |
| − | |Nekrose||Thrombose
| |
| − | |-
| |
| − | |Nekrose haemorrhagisch||Ja
| |
| − | |-
| |
| − | |Nekrose Infarkt||Ja
| |
| − | |-
| |
| − | |Nephrostomie||Moeglich
| |
| − | |-
| |
| − | |NHSBSP||B1a
| |
| − | |-
| |
| − | |NHSBSP||B1b
| |
| − | |-
| |
| − | |NHSBSP||B2
| |
| − | |-
| |
| − | |NHSBSP||B3
| |
| − | |-
| |
| − | |NHSBSP||B4
| |
| − | |-
| |
| − | |NHSBSP||B5a
| |
| − | |-
| |
| − | |NHSBSP||B5b
| |
| − | |-
| |
| − | |NHSBSP||B5c
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B1a
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B1b
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B2
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B3
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B4
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B5a
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B5b
| |
| − | |-
| |
| − | |NHSBSP-Schnellschnitt||B5c
| |
| − | |-
| |
| − | |Niere||fragmentiert
| |
| − | |-
| |
| − | |Niere||intakt
| |
| − | |-
| |
| − | |Niere||Lithiasis ja
| |
| − | |-
| |
| − | |Niere||Lithiasis klinisch ja
| |
| − | |-
| |
| − | |Niere||Lithiasis nein
| |
| − | |-
| |
| − | |Niere||zerklueftet
| |
| − | |-
| |
| − | |Nierendegeneration polycystisch adult||Moeglich
| |
| − | |-
| |
| − | |Oedem||Chronisch
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis Crohn klinisch Verdacht
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis eosinophil moeglich
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis infektioes Pilz
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis infektioes Vir
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis infektioes Vir CMV
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis infektioes Vir Herpes
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis radiogen moeglich
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung Oesophagitis unklassifiziert
| |
| − | |-
| |
| − | |Oesophagus||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis Grad 1
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis Grad 2
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis Grad 3
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis Grad 4
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis Grad unbekannt
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis unbekannt
| |
| − | |-
| |
| − | |Oesophagus||Klinisch Refluxoesophagitis Verdacht
| |
| − | |-
| |
| − | |Oesophagus||Regelhaft
| |
| − | |-
| |
| − | |Ossfication metaplastisch||Ja
| |
| − | |-
| |
| − | |Ossification||Ja
| |
| − | |-
| |
| − | |Ossification metaplastisch||Moeglich
| |
| − | |-
| |
| − | |Ossifikation metaplastisch||Ja
| |
| − | |-
| |
| − | |Otosklerose||Moeglich
| |
| − | |-
| |
| − | |Papillaere Laesion||Ja
| |
| − | |-
| |
| − | |Papillom||Ja
| |
| − | |-
| |
| − | |Papillom||Moeglich
| |
| − | |-
| |
| − | |Parasit||Helminth
| |
| − | |-
| |
| − | |Parasit||Unklassifiziert
| |
| − | |-
| |
| − | |Perforation||Klinisch
| |
| − | |-
| |
| − | |Perforation ||Klinisch iatrogen
| |
| − | |-
| |
| − | |Perforation ||Moeglich
| |
| − | |-
| |
| − | |Perforation klinisch||Ja
| |
| − | |-
| |
| − | |Photodynamische Diagnostik PDD||Negativ
| |
| − | |-
| |
| − | |Photodynamische Diagnostik PDD||Positiv
| |
| − | |-
| |
| − | |Phylloider Tumor||Moeglich
| |
| − | |-
| |
| − | |Portio||Intakt
| |
| − | |-
| |
| − | |Portio||Zerklüftet
| |
| − | |-
| |
| − | |Position||unbekannt
| |
| − | |-
| |
| − | |Praeparat||Fragmentiert
| |
| − | |-
| |
| − | |Praeparat||Intakt
| |
| − | |-
| |
| − | |Praeparat||Zerklueftet
| |
| − | |-
| |
| − | |Praeputium||Entzuendung akut Abszess
| |
| − | |-
| |
| − | |Praeputium||Entzuendung akut unklassifiziert
| |
| − | |-
| |
| − | |Praeputium||Entzuendung granulomatoes
| |
| − | |-
| |
| − | |Praeputium||Entzuendung infektioes Pilz
| |
| − | |-
| |
| − | |Praeputium||Entzuendung Lichen ruber planus moeglich
| |
| − | |-
| |
| − | |Praeputium||Entzuendung Lichen sclerosus et atrophicus
| |
| − | |-
| |
| − | |Praeputium||Entzuendung Nekrose fibrinoid
| |
| − | |-
| |
| − | |Praeputium||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Praeputium||Ulcus
| |
| − | |-
| |
| − | |Präparatradiogramm||Ja
| |
| − | |-
| |
| − | |Präparatradiogramm||Kalk ja
| |
| − | |-
| |
| − | |Präparatradiogramm||Kalk moeglich
| |
| − | |-
| |
| − | |Präparatradiogramm||Kalk nein
| |
| − | |-
| |
| − | |Präparatradiogramm||Kalk unklar
| |
| − | |-
| |
| − | |Präparatradiogramm||Nein
| |
| − | |-
| |
| − | |Präparatradiogramm||Unbekannt
| |
| − | |-
| |
| − | |Proliferation fibromuskulaer||Ja
| |
| − | |-
| |
| − | |Prostata||Entzuendung granulomatoes
| |
| − | |-
| |
| − | |Prostata||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Prostata||Entzuendung unklassifiziert akut
| |
| − | |-
| |
| − | |Prostata||Entzuendung unklassifiziert chronisch
| |
| − | |-
| |
| − | |Prostata||Entzuendung unklassifiziert chronisch floride
| |
| − | |-
| |
| − | |Prostata||fragmentiert
| |
| − | |-
| |
| − | |Prostata||intakt
| |
| − | |-
| |
| − | |Prostata||zerklueftet
| |
| − | |-
| |
| − | |PSA||unbekannt
| |
| − | |-
| |
| − | |Pseudarthrose||Moeglich
| |
| − | |-
| |
| − | |R0||Resektat quintaer
| |
| − | |-
| |
| − | |R0||Resektat sekundaer
| |
| − | |-
| |
| − | |R1||Aboral
| |
| − | |-
| |
| − | |R1||Anterior (hautwaerts)
| |
| − | |-
| |
| − | |R1||Apex
| |
| − | |-
| |
| − | |R1||Apex links
| |
| − | |-
| |
| − | |R1||Apex rechts
| |
| − | |-
| |
| − | |R1||Basis links
| |
| − | |-
| |
| − | |R1||Basis rechts
| |
| − | |-
| |
| − | |R1||Circumferenz
| |
| − | |-
| |
| − | |R1||Circumferenz extraperitoneal
| |
| − | |-
| |
| − | |R1||Kaudal
| |
| − | |-
| |
| − | |R1||Kranial
| |
| − | |-
| |
| − | |R1||Lateral
| |
| − | |-
| |
| − | |R1||Lateral links
| |
| − | |-
| |
| − | |R1||Lateral rechts
| |
| − | |-
| |
| − | |R1||Mamillaer
| |
| − | |-
| |
| − | |R1||Medial
| |
| − | |-
| |
| − | |R1||Mesocolisch
| |
| − | |-
| |
| − | |R1||Mesorectal
| |
| − | |-
| |
| − | |R1||Multiloculaer
| |
| − | |-
| |
| − | |R1||Nachresektat
| |
| − | |-
| |
| − | |R1||Oral
| |
| − | |-
| |
| − | |R1||Pankreasschwanz
| |
| − | |-
| |
| − | |R1||Peripher
| |
| − | |-
| |
| − | |R1||Posterior (pectoraliswaerts)
| |
| − | |-
| |
| − | |R1||Posterior links
| |
| − | |-
| |
| − | |R1||Posterior rechts
| |
| − | |-
| |
| − | |R1||Resektat quartaer
| |
| − | |-
| |
| − | |R1||Resektat sekundaer
| |
| − | |-
| |
| − | |R1||Samenblase links
| |
| − | |-
| |
| − | |R1||Samenblase rechts
| |
| − | |-
| |
| − | |R1||Unbezeichnet
| |
| − | |-
| |
| − | |R1||Ventral links
| |
| − | |-
| |
| − | |R1||Ventral rechts
| |
| − | |-
| |
| − | |R2||Aboral
| |
| − | |-
| |
| − | |R2||Circumferenz
| |
| − | |-
| |
| − | |R2||Mesocolisch
| |
| − | |-
| |
| − | |Regression||Dworak
| |
| − | |-
| |
| − | |Reif||Ja
| |
| − | |-
| |
| − | |Reifezustand||Reif
| |
| − | |-
| |
| − | |Reifezustand||Unreif
| |
| − | |-
| |
| − | |Reifezustand||Vorgereift
| |
| − | |-
| |
| − | |Reinke Oedem||Moeglich
| |
| − | |-
| |
| − | |Resektat||negativ
| |
| − | |-
| |
| − | |Resektat||positiv
| |
| − | |-
| |
| − | |Resektat||unklar
| |
| − | |-
| |
| − | |Resektat multipel||positiv
| |
| − | |-
| |
| − | |Resektat quartaer||negativ
| |
| − | |-
| |
| − | |Resektat quartaer||positiv
| |
| − | |-
| |
| − | |Resektat quartaer||unklar
| |
| − | |-
| |
| − | |Resektat quintaer||positiv
| |
| − | |-
| |
| − | |Resektat quintaer||unklar
| |
| − | |-
| |
| − | |Resektat sekundaer||negativ
| |
| − | |-
| |
| − | |Resektat sekundaer||positiv
| |
| − | |-
| |
| − | |Resektat sekundaer||Stanze
| |
| − | |-
| |
| − | |Resektat sekundaer||unklar
| |
| − | |-
| |
| − | |Resektat tertiaer||negativ
| |
| − | |-
| |
| − | |Resektat tertiaer||positiv
| |
| − | |-
| |
| − | |Reservezellen||Ja
| |
| − | |-
| |
| − | |Reservezellen||Nein
| |
| − | |-
| |
| − | |Risiko||Hoch
| |
| − | |-
| |
| − | |Risiko||Niedrig
| |
| − | |-
| |
| − | |RR min||Aboral
| |
| − | |-
| |
| − | |RR min||Anterior
| |
| − | |-
| |
| − | |RR min||Anterior (hautwaerts)
| |
| − | |-
| |
| − | |RR min||Anterior extraperitoneal
| |
| − | |-
| |
| − | |RR min||Apex links
| |
| − | |-
| |
| − | |RR min||Apex rechts
| |
| − | |-
| |
| − | |RR min||Basis links
| |
| − | |-
| |
| − | |RR min||Basis rechts
| |
| − | |-
| |
| − | |RR min||Circumferenz
| |
| − | |-
| |
| − | |RR min||Corpuswaerts
| |
| − | |-
| |
| − | |RR min||Dorsal links
| |
| − | |-
| |
| − | |RR min||Dorsal rechts
| |
| − | |-
| |
| − | |RR min||Kaudal
| |
| − | |-
| |
| − | |RR min||Kranial
| |
| − | |-
| |
| − | |RR min||Lateral
| |
| − | |-
| |
| − | |RR min||Lateral links
| |
| − | |-
| |
| − | |RR min||Lateral rechts
| |
| − | |-
| |
| − | |RR min||Links
| |
| − | |-
| |
| − | |RR min||Mamillaer
| |
| − | |-
| |
| − | |RR min||Medial
| |
| − | |-
| |
| − | |RR min||Mesorectal
| |
| − | |-
| |
| − | |RR min||Oral
| |
| − | |-
| |
| − | |RR min||Parametrien links
| |
| − | |-
| |
| − | |RR min||Peripher
| |
| − | |-
| |
| − | |RR min||Posterior
| |
| − | |-
| |
| − | |RR min||Posterior (pectoraliswaerts)
| |
| − | |-
| |
| − | |RR min||Posterior extraperitoneal
| |
| − | |-
| |
| − | |RR min||Rechts
| |
| − | |-
| |
| − | |RR min||Serosawaerts
| |
| − | |-
| |
| − | |RR min||Unbezeichnet
| |
| − | |-
| |
| − | |RR min||Vaginalwaerts
| |
| − | |-
| |
| − | |RR min||Ventral links
| |
| − | |-
| |
| − | |RR min||Ventral rechts
| |
| − | |-
| |
| − | |Ruptur||Moeglich
| |
| − | |-
| |
| − | |Rx||Aboral
| |
| − | |-
| |
| − | |Rx||Anterior (hautwaerts)
| |
| − | |-
| |
| − | |Rx||Anterior extraperitoneal
| |
| − | |-
| |
| − | |Rx||Apex
| |
| − | |-
| |
| − | |Rx||Apex links
| |
| − | |-
| |
| − | |Rx||Apex rechts
| |
| − | |-
| |
| − | |Rx||Basis
| |
| − | |-
| |
| − | |Rx||Basis links
| |
| − | |-
| |
| − | |Rx||Basis rechts
| |
| − | |-
| |
| − | |Rx||Caudal
| |
| − | |-
| |
| − | |Rx||Circumferenz
| |
| − | |-
| |
| − | |Rx||Dorsal
| |
| − | |-
| |
| − | |Rx||Kaudal
| |
| − | |-
| |
| − | |Rx||Kranial
| |
| − | |-
| |
| − | |Rx||Lateral
| |
| − | |-
| |
| − | |Rx||Lateral links
| |
| − | |-
| |
| − | |Rx||Lateral rechts
| |
| − | |-
| |
| − | |Rx||Mamillaer
| |
| − | |-
| |
| − | |Rx||Medial
| |
| − | |-
| |
| − | |Rx||Nachresektat
| |
| − | |-
| |
| − | |Rx||Peripher
| |
| − | |-
| |
| − | |Rx||Posterior (pectoraliswaerts)
| |
| − | |-
| |
| − | |Rx||Posterior links
| |
| − | |-
| |
| − | |Rx||Posterior rechts
| |
| − | |-
| |
| − | |Rx||Resektat multipel
| |
| − | |-
| |
| − | |Rx||Resektat sekundaer
| |
| − | |-
| |
| − | |Rx||Resektat tertiaer
| |
| − | |-
| |
| − | |Rx||Samenblase rechts
| |
| − | |-
| |
| − | |Rx||Unbezeichnet
| |
| − | |-
| |
| − | |Rx||Ventral
| |
| − | |-
| |
| − | |Rx||Ventral links
| |
| − | |-
| |
| − | |Rx||Ventral rechts
| |
| − | |-
| |
| − | |Schilddruese||Entzuendung granulomatoes
| |
| − | |-
| |
| − | |Schilddruese||Entzuendung granulomatoes de Quervain
| |
| − | |-
| |
| − | |Schilddruese||Entzuendung infektioes
| |
| − | |-
| |
| − | |Schilddruese||Entzuendung infektioes klinisch
| |
| − | |-
| |
| − | |Schilddruese||Entzuendung lymphozytaer Hashimoto
| |
| − | |-
| |
| − | |Schilddruese||Entzuendung lymphozytaer unklassifiziert
| |
| − | |-
| |
| − | |Schilddruese||Klinisch Basedow
| |
| − | |-
| |
| − | |Schleimhaut||Gekappt
| |
| − | |-
| |
| − | |Schleimhaut||Vollstaendig
| |
| − | |-
| |
| − | |Schnellschnitt||positiv
| |
| − | |-
| |
| − | |Speicheldruese||Entzuendung unklassifiziert
| |
| − | |-
| |
| − | |Spermatocele||Ja
| |
| − | |-
| |
| − | |Spermatocele||Moeglich
| |
| − | |-
| |
| − | |Spermatocele Rete testis||Ja
| |
| − | |-
| |
| − | |Sprue||Kein Anhalt
| |
| − | |-
| |
| − | |Sprue||moeglich
| |
| − | |-
| |
| − | |Stanzbiopsie||Bekannt
| |
| − | |-
| |
| − | |Stanzbiopsie||Unbekannt
| |
| − | |-
| |
| − | |Stanze Qualitaet||ausreichend
| |
| − | |-
| |
| − | |Stanze Qualitaet||gut
| |
| − | |-
| |
| − | |Stanze Qualitaet||unzureichend
| |
| − | |-
| |
| − | |Therapie||Unbekannt
| |
| − | |-
| |
| − | |Therapie BCG||Ja
| |
| − | |-
| |
| − | |Therapie BCG||Nein
| |
| − | |-
| |
| − | |Therapie BCG||Unbekannt
| |
| − | |-
| |
| − | |Therapie Bestrahlung||Ja
| |
| − | |-
| |
| − | |Therapie Bestrahlung||Nein
| |
| − | |-
| |
| − | |Therapie Bestrahlung||Unbekannt
| |
| − | |-
| |
| − | |Therapie Chemotherapie||Ja
| |
| − | |-
| |
| − | |Therapie Chemotherapie||Nein
| |
| − | |-
| |
| − | |Therapie Chemotherapie||Unbekannt
| |
| − | |-
| |
| − | |Therapie endokrin||ja
| |
| − | |-
| |
| − | |Therapie endokrin||nein
| |
| − | |-
| |
| − | |Therapie endokrin||unbekannt
| |
| − | |-
| |
| − | |Therapie Hormonentzug||Ja
| |
| − | |-
| |
| − | |Therapie Laser||Ja
| |
| − | |-
| |
| − | |Therapie Mitomycin||Ja
| |
| − | |-
| |
| − | |Therapie Mitomycin||Unbekannt
| |
| − | |-
| |
| − | |Therapie Priming||Ja
| |
| − | |-
| |
| − | |Therapie Radiatio||ja
| |
| − | |-
| |
| − | |Therapie Radiatio||nein
| |
| − | |-
| |
| − | |Therapie Radiatio||unbekannt
| |
| − | |-
| |
| − | |Therapie Steroide||Ja
| |
| − | |-
| |
| − | |Thrombus||Aelter
| |
| − | |-
| |
| − | |Thrombus||Eisen ja
| |
| − | |-
| |
| − | |Thrombus||Eisen nein
| |
| − | |-
| |
| − | |Thrombus||Frisch
| |
| − | |-
| |
| − | |Thrombus||Frischer
| |
| − | |-
| |
| − | |Thrombus||Organisat unvollstaendig
| |
| − | |-
| |
| − | |Thrombus||Organisat vollstaendig
| |
| − | |-
| |
| − | |Thrombus intervilloes||Ja
| |
| − | |-
| |
| − | |Thrombus intervilloes||Nein
| |
| − | |-
| |
| − | |Torsion klinisch||Ja
| |
| − | |-
| |
| − | |Torsion klinisch||Moeglich
| |
| − | |-
| |
| − | |Torsion klinisch||Unbekannt
| |
| − | |-
| |
| − | |Transformation progressiv||Ja
| |
| − | |-
| |
| − | |Transformation progressiv||Nein
| |
| − | |-
| |
| − | |Transformation progressiv||Unklar
| |
| − | |-
| |
| − | |Transurethrale Resektion Zustand nach||Ja
| |
| − | |-
| |
| − | |Trichomonaden||Nein
| |
| − | |-
| |
| − | |Trophoblastreaktion ueberschiessend||Ja
| |
| − | |-
| |
| − | |Tumor||fragmentiert
| |
| − | |-
| |
| − | |Tumor||Intact
| |
| − | |-
| |
| − | |Tumor||intakt
| |
| − | |-
| |
| − | |Tumor||metachron
| |
| − | |-
| |
| − | |Tumor||Ruptur
| |
| − | |-
| |
| − | |Tumor||synchron
| |
| − | |-
| |
| − | |Tumor||zerklueftet
| |
| − | |-
| |
| − | |Tumor Durchmesser||Unklar
| |
| − | |-
| |
| − | |Tumor Fettgewebe||ja
| |
| − | |-
| |
| − | |Tumor fragmentiert||Ja
| |
| − | |-
| |
| − | |Tumor inflammatorisch myofibroblastisch||Ja
| |
| − | |-
| |
| − | |Tumor intakt||Ja
| |
| − | |-
| |
| − | |Tumor intakt||Nein
| |
| − | |-
| |
| − | |Tumor Karzinom lymphoepithelial aehnlich||Ja
| |
| − | |-
| |
| − | |Tumor Karzinom plasmacytoid||Ja
| |
| − | |-
| |
| − | |Tumor Karzinom Riesenzelle trophoblastaer||Ja
| |
| − | |-
| |
| − | |Tumor multifokal||Metachron
| |
| − | |-
| |
| − | |Tumor multifokal||Synchron
| |
| − | |-
| |
| − | |Tumor perineural||ja
| |
| − | |-
| |
| − | |Tumor perineural||nein
| |
| − | |-
| |
| − | |Tumor perineural||unbekannt
| |
| − | |-
| |
| − | |Tumor ulceriert||ja
| |
| − | |-
| |
| − | |Tumor ulceriert||nein
| |
| − | |-
| |
| − | |Tumor Volumen||Unklar
| |
| − | |-
| |
| − | |Tumorausbreitung||Mesenteriolum
| |
| − | |-
| |
| − | |Tumorausbreitung||Muskularis propria
| |
| − | |-
| |
| − | |Tumorausbreitung||Muskulatur
| |
| − | |-
| |
| − | |Tumorausbreitung||Submukosa
| |
| − | |-
| |
| − | |Tumorausbreitung||Submukosa mittleres Drittel
| |
| − | |-
| |
| − | |Tumorausbreitung||Submukosa oberes Drittel
| |
| − | |-
| |
| − | |Tumorausbreitung||Submukosa unteres Drittel
| |
| − | |-
| |
| − | |Tumordurchmesser||Unbekannt
| |
| − | |-
| |
| − | |Tumorposition||dorsal
| |
| − | |-
| |
| − | |Tumorposition||Extraperitoneal
| |
| − | |-
| |
| − | |Tumorposition||Intraperitoneal
| |
| − | |-
| |
| − | |Tumorposition||Intraperitoneal Extraperitoneal
| |
| − | |-
| |
| − | |Tumorposition||links
| |
| − | |-
| |
| − | |Tumorposition||rechts
| |
| − | |-
| |
| − | |Tumorposition||unklar
| |
| − | |-
| |
| − | |Tumorposition||ventral
| |
| − | |-
| |
| − | |Tumorwachstum||dissoziiert
| |
| − | |-
| |
| − | |Tumorwachstum||solide
| |
| − | |-
| |
| − | |Typ||1
| |
| − | |-
| |
| − | |Typ||2
| |
| − | |-
| |
| − | |Typ||Azinaer
| |
| − | |-
| |
| − | |Typ||Ductal
| |
| − | |-
| |
| − | |Typ||Klarzellig
| |
| − | |-
| |
| − | |Typ||Microacinaer
| |
| − | |-
| |
| − | |Typ||Mucinoes
| |
| − | |-
| |
| − | |Typ||Multiloculaer cystisch
| |
| − | |-
| |
| − | |Typ||Schaumzellig
| |
| − | |-
| |
| − | |Typ||Unbestimmt
| |
| − | |-
| |
| − | |Typ||Unklassifiziert mucinoes spindelzellig
| |
| − | |-
| |
| − | |Unreif||Ja
| |
| − | |-
| |
| − | |Untersuchung||Klinisch nicht erwuenscht
| |
| − | |-
| |
| − | |Ureter||intakt
| |
| − | |-
| |
| − | |Ureter||zerklueftet
| |
| − | |-
| |
| − | |Varicocele||Moeglich
| |
| − | |-
| |
| − | |Varicosis||Ja
| |
| − | |-
| |
| − | |Varicosis||Klinisch
| |
| − | |-
| |
| − | |Varicosis||Klinisch Verdacht
| |
| − | |-
| |
| − | |Varicosis||Moeglich
| |
| − | |-
| |
| − | |Vaskulitis||Ja
| |
| − | |-
| |
| − | |Vaskulitis||Moeglich
| |
| − | |-
| |
| − | |Velamentoeser Nabelschnuransatz||Ja
| |
| − | |-
| |
| − | |Volumen||Unklar
| |
| − | |-
| |
| − | |Volumen klinisch||unbekannt
| |
| − | |-
| |
| − | |Wachstumsmuster cribriform||Ja
| |
| − | |-
| |
| − | |Wachstumsmuster gemischt solide cribriform||Ja
| |
| − | |-
| |
| − | |Wachstumsmuster gemischt solide papillaer||Ja
| |
| − | |-
| |
| − | |Wachstumsmuster lobulaer||Ja
| |
| − | |-
| |
| − | |Wachstumsmuster papillaer||Ja
| |
| − | |-
| |
| − | |Wachstumsmuster solide||Ja
| |
| − | |-
| |
| − | |Zottenreifungsstoerung||Ja
| |
| − | |-
| |
| − | |Zottenreifungsstoerung||Nein
| |
| − | |-
| |
| − | |Zottenstroma Fibrose||Ja
| |
| − | |-
| |
| − | |Zwillingsplazenta||Dichorial diamnial fusioniert
| |
| − | |-
| |
| − | |Zwillingsplazenta||Dichorial diamnial getrennt
| |
| − | |-
| |
| − | |Zwillingsplazenta||Monochorial diamnial
| |
| − | |-
| |
| − | |Zwillingsplazenta||Monochorial monoamnial
| |
| − | |-
| |
| − | |Zwillingsplazenta||Transfusionsysndrom
| |
| − | |-
| |
| − | |Zwillingsplazenta||Typ nicht bestimmbar
| |
| − | |-
| |
| − | |Zyklustermin||Unbekannt
| |
| − | |-
| |
| − | |Zylinderzellhyperplasie||Ja
| |
| − | |-
| |
| − | |Zylinderzellmetaplasie||Ja
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 22: Attribut-Code-Paare (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ==65. Attribute-Werte-Paare== | |
| − | | |
| − | {{AlertBox|Die folgende Tabelle muss auf jeden Fall hinsichtlich der Werte noch aufgeräumt werden!
| |
| − | Die Werte werden als Datentyp PQ übermittelt. Wenn es sich um relative Angaben handelt (">"), dann sollte IVL verwendet werden.
| |
| − | Einige Werte sollten besser als Codes dargestellt werden. Diese sind in rot markiert.
| |
| | }} | | }} |
| | | | |
| − | {| class="hl7table" | + | {{HL7transclude|cdapath:Einleitung}} |
| − | !Code!!Codename!!Bedeutung!!Einheiten
| + | {{HL7transclude|cdapath:Vol1Profiles}} |
| − | |-
| + | {{HL7transclude|cdapath:Vol3ContentModules}} |
| − | |||Abstand aboral||||mm
| |
| − | |-
| |
| − | |||Abstand anterior extraperitoneal||||mm
| |
| − | |-
| |
| − | |||Abstand anterior serosawaerts||||mm
| |
| − | |-
| |
| − | |||Abstand caudal||||mm
| |
| − | |-
| |
| − | |||Abstand Circumferenz||||mm
| |
| − | |-
| |
| − | |||Abstand cranial||||mm
| |
| − | |-
| |
| − | |||Abstand dorsal||||mm
| |
| − | |-
| |
| − | |||Abstand Knoten Basis||||mm
| |
| − | |-
| |
| − | |||Abstand Knoten Basis (weniger als)||||mm
| |
| − | |-
| |
| − | |||Abstand lateral ()||||mm
| |
| − | |-
| |
| − | |||Abstand milzwaerts||||mm
| |
| − | |-
| |
| − | |||Abstand oral||||mm
| |
| − | |-
| |
| − | |||Abstand papillenwaerts||||mm
| |
| − | |-
| |
| − | |||Abstand posterior||||mm
| |
| − | |-
| |
| − | |||Abstand posterior extraperitoneal||||mm
| |
| − | |-
| |
| − | |||Abstand serosawaerts||||mm
| |
| − | |-
| |
| − | |||Abstand unbezeichnet||||mm
| |
| − | |-
| |
| − | |||Abstand ventral||||mm
| |
| − | |-
| |
| − | |||Anteil intraductal||||%
| |
| − | |-
| |
| − | |||Anteil muzinoes||||%
| |
| − | |-
| |
| − | |||Anzahl||||
| |
| − | |-
| |
| − | |||Anzahl mehr als||||
| |
| − | |-
| |
| − | |||Anzahl Praeparate||||
| |
| − | |-
| |
| − | |||Apex links||||
| |
| − | |-
| |
| − | |||Atrophie Antrum||||
| |
| − | |-
| |
| − | |||Atrophie Corpus||||
| |
| − | |-
| |
| − | |||Atrophie Magen||||
| |
| − | |-
| |
| − | |||Basis rechts||||
| |
| − | |-
| |
| − | |||Beurteilbare Markraeume||||
| |
| − | |-
| |
| − | |||Chorionkarzinom ||||%
| |
| − | |-
| |
| − | |||cISH Signale je Zellkern <||||
| |
| − | |-
| |
| − | |||cISH Signale je Zellkern >||||
| |
| − | |-
| |
| − | |||Clark Level||||
| |
| − | |-
| |
| − | |||Dicke||||mm
| |
| − | |-
| |
| − | |||Dottersacktumor ||||%
| |
| − | |-
| |
| − | |||Durchmesser||||mm
| |
| − | |-
| |
| − | |||Durchmesser >||||mm
| |
| − | |-
| |
| − | |||Durchmesser groesser||||mm
| |
| − | |-
| |
| − | |||Durchmesser kleiner||||mm
| |
| − | |-
| |
| − | |||Durchmesser 1||||mm
| |
| − | |-
| |
| − | |||Durchmesser 2||||mm
| |
| − | |-
| |
| − | |||Durchmesser klinisch||||mm
| |
| − | |-
| |
| − | |||Durchmesser laengs||||mm
| |
| − | |-
| |
| − | |||Durchmesser Metastase||||mm
| |
| − | |-
| |
| − | |||Durchmesser quer||||mm
| |
| − | |-
| |
| − | |||Embryonales Karzinom ||||%
| |
| − | |-
| |
| − | |||Entzuendung Aktivitaet||||
| |
| − | |-
| |
| − | |||Entzuendung geringgradig||||
| |
| − | |-
| |
| − | |||Entzuendung minimal||||
| |
| − | |-
| |
| − | |||Entzuendung mittelgradig||||
| |
| − | |-
| |
| − | |||Entzündungsgrad||||
| |
| − | |-
| |
| − | |||Eosinophile starke Vergroesserung mehr als||||
| |
| − | |-
| |
| − | |||Fibrose Dicke||||mm
| |
| − | |-
| |
| − | |||Fibrose geringgradig||||
| |
| − | |-
| |
| − | |||Fibrose mittelgradig||||
| |
| − | |-
| |
| − | |||Fibrose nein||||
| |
| − | |-
| |
| − | |||Fibrose schwergradig||||
| |
| − | |-
| |
| − | |||Fibrose Zirrhose||||
| |
| − | |-
| |
| − | |||Gewicht||||g
| |
| − | |-
| |
| − | |||Gewicht Praeparat||||g
| |
| − | |-
| |
| − | |||Gewicht Praeparat weniger als||||g
| |
| − | |-
| |
| − | |||Gleason 2||||%
| |
| − | |-
| |
| − | |||Gleason 3||||%
| |
| − | |-
| |
| − | |||Gleason 4||||%
| |
| − | |-
| |
| − | |||Gleason 5||||%
| |
| − | |-
| |
| − | |||Gleason tertiaer||||
| |
| − | |-
| |
| − | |||Herd klinisch||||
| |
| − | |-
| |
| − | |||High grade||||
| |
| − | |-
| |
| − | |||High risk||||
| |
| − | |-
| |
| − | |||Intermediate grade||||
| |
| − | |-
| |
| − | |||Intraduktaler Anteil ||||%
| |
| − | |-
| |
| − | |||Intraepitheliale Neoplasie High Grade||||
| |
| − | |-
| |
| − | |||Intraepitheliale Neoplasie Low Grade||||
| |
| − | |-
| |
| − | |||Kalk||||mm
| |
| − | |-
| |
| − | |||Kalkfeld histologisch||||mm
| |
| − | |-
| |
| − | |||Kalkfeld radiologisch||||mm
| |
| − | |-
| |
| − | |||Lobulaere Intraepitheliale Neoplasie||||
| |
| − | |-
| |
| − | |||Low grade||||
| |
| − | |-
| |
| − | |||Low malignant potential||||
| |
| − | |-
| |
| − | |||Low risk||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe 01||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe 02||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe 03||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe 04||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe 05||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe 08||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links 01||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links 02||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links 03||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links 04||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links 05||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links 06||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe links mehrere||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe mehrere||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts 01||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts 02||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts 03||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts 04||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts 05||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts 06||||
| |
| − | |-
| |
| − | |||Lymphknoten positiv Gruppe rechts mehrere||||
| |
| − | |-
| |
| − | |||Malignitaetsgrad nucleaer||||
| |
| − | |-
| |
| − | |||Mastzellen||||qmm
| |
| − | |-
| |
| − | |||Mikrofibrin Grad||||
| |
| − | |-
| |
| − | |||Nebenschilddruese||||
| |
| − | |-
| |
| − | |||Nebenschilddruese links||||
| |
| − | |-
| |
| − | |||Nebenschilddruese rechts||||
| |
| − | |-
| |
| − | |||NULL||||
| |
| − | |-
| |
| − | |||Objekttraeger||||
| |
| − | |-
| |
| − | |||Portalfelder||||
| |
| − | |-
| |
| − | |||Position (Zifferblatt)||||
| |
| − | |-
| |
| − | |||Position klinisch||||cm
| |
| − | |-
| |
| − | |||Position Mammotomschema A||||
| |
| − | |-
| |
| − | |||Position Mammotomschema A Kalk||||
| |
| − | |-
| |
| − | |||Position Mammotomschema B||||
| |
| − | |-
| |
| − | |||Position Mammotomschema B Kalk||||
| |
| − | |-
| |
| − | |||Position Mammotomschema C||||
| |
| − | |-
| |
| − | |||Position Mammotomschema C Kalk||||
| |
| − | |-
| |
| − | |||Position Mammotomschema D||||
| |
| − | |-
| |
| − | |||Position Mammotomschema D Kalk||||
| |
| − | |-
| |
| − | |||Position Mammotomschema E Kalk||||
| |
| − | |-
| |
| − | |||Probe||||
| |
| − | |-
| |
| − | |||Proliferationsgrad Schmitt||||
| |
| − | |-
| |
| − | |||PSA||||ng/ml
| |
| − | |-
| |
| − | |||QM Score gering||||
| |
| − | |-
| |
| − | |||QM Score optimal||||
| |
| − | |-
| |
| − | |||QM Score suboptimal||||
| |
| − | |-
| |
| − | |||Regressionsgrad||||
| |
| − | |-
| |
| − | |||Regressionsgrad (Sinn)||||
| |
| − | |-
| |
| − | |||Resektat multipel Nummer||||
| |
| − | |-
| |
| − | |||RRmin anterior (hautwaerts)||||mm
| |
| − | |-
| |
| − | |||RRmin anterior (hautwaerts) groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin anterior (hautwaerts) kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin Apex links||||mm
| |
| − | |-
| |
| − | |||RRmin Apex rechts||||mm
| |
| − | |-
| |
| − | |||RRmin Basis||||mm
| |
| − | |-
| |
| − | |||RRmin Basis links||||mm
| |
| − | |-
| |
| − | |||RRmin Basis rechts||||mm
| |
| − | |-
| |
| − | |||RRmin groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin kaudal||||mm
| |
| − | |-
| |
| − | |||RRmin kaudal groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin kaudal kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin kranial||||mm
| |
| − | |-
| |
| − | |||RRmin kranial groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin kranial kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin lateral||||mm
| |
| − | |-
| |
| − | |||RRmin lateral groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin lateral kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin Lateral links||||mm
| |
| − | |-
| |
| − | |||RRmin Lateral rechts||||mm
| |
| − | |-
| |
| − | |||RRmin mamillaer||||mm
| |
| − | |-
| |
| − | |||RRmin mamillaer groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin mamillaer kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin medial||||mm
| |
| − | |-
| |
| − | |||RRmin medial groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin medial kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin peripher||||mm
| |
| − | |-
| |
| − | |||RRmin peripher groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin peripher kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin posterior (pectoraliswaerts)||||mm
| |
| − | |-
| |
| − | |||RRmin posterior groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin posterior kleiner als||||mm
| |
| − | |-
| |
| − | |||RRmin posterior links||||mm
| |
| − | |-
| |
| − | |||RRmin posterior rechts||||mm
| |
| − | |-
| |
| − | |||RRmin Samenblase rechts||||mm
| |
| − | |-
| |
| − | |||RRmin unbezeichnet||||mm
| |
| − | |-
| |
| − | |||RRmin unbezeichnet groesser als||||mm
| |
| − | |-
| |
| − | |||RRmin Ventral||||mm
| |
| − | |-
| |
| − | |||Ruptur klinisch vor ||||d
| |
| − | |-
| |
| − | |||Schnitte||||
| |
| − | |-
| |
| − | |||Schwangerschaftswoche||||
| |
| − | |-
| |
| − | |||Score Elston Ellis||||
| |
| − | |-
| |
| − | |||Seminom ||||%
| |
| − | |-
| |
| − | |||Seminom spermatozytisch ||||%
| |
| − | |-
| |
| − | |||Spaene positiv||||%
| |
| − | |-
| |
| − | |||Spermatogonien 50 Tubuli||||
| |
| − | |-
| |
| − | |||Stanzzylinder||||
| |
| − | |-
| |
| − | |||Stanzzylinder positiv||||
| |
| − | |-
| |
| − | |||Stanzzylinder positiv links||||
| |
| − | |-
| |
| − | |||Stanzzylinder positiv Mitte||||
| |
| − | |-
| |
| − | |||Stanzzylinder positiv rechts||||
| |
| − | |-
| |
| − | |||STUMP||||
| |
| − | |-
| |
| − | |||Teratom ||||%
| |
| − | |-
| |
| − | |||Torsion klinisch seit||||h
| |
| − | |-
| |
| − | |||Tubuli Spermatogenese Spermatiden erhalten ||||%
| |
| − | |-
| |
| − | |||Tubuli Spermatogenese Spermatogonien erhalten ||||%
| |
| − | |-
| |
| − | |||Tubuli Spermatogenese Spermatozoen erhalten ||||%
| |
| − | |-
| |
| − | |||Tubuli Spermatogenese Spermatozyten erhalten ||||%
| |
| − | |-
| |
| − | |||Tubulus Durchmesser||||mm
| |
| − | |-
| |
| − | |||Tumor||||
| |
| − | |-
| |
| − | |||Tumor Anteil||||%
| |
| − | |-
| |
| − | |||Tumor Anteil kleiner als||||%
| |
| − | |-
| |
| − | |||Tumor Invasionstiefe||||mm
| |
| − | |-
| |
| − | |||Tumor multifokal Herde||||
| |
| − | |-
| |
| − | |||Tumor multifokal Herde mehr als||||
| |
| − | |-
| |
| − | |||Tumordicke||||mm
| |
| − | |-
| |
| − | |||Tumorgewicht ||||g
| |
| − | |-
| |
| − | |||Tumorvolumen||||ml
| |
| − | |-
| |
| − | |||Unreifer Anteil ||||%
| |
| − | |-
| |
| − | |||Unreifer Anteil kleiner ||||%
| |
| − | |-
| |
| − | |||Van Nuys Index||||
| |
| − | |-
| |
| − | |||Verfettung ||||%
| |
| − | |-
| |
| − | |||Vitaler Anteil ||||%
| |
| − | |-
| |
| − | |||Vitaler Anteil groesser ||||%
| |
| − | |-
| |
| − | |||Vitaler Anteil kleiner ||||%
| |
| − | |-
| |
| − | |||Volumen klinisch||||ml
| |
| − | |-
| |
| − | |||Wanddicke||||mm
| |
| − | |-
| |
| − | |||Wotherspoon||||
| |
| − | |-
| |
| − | |||Zahl||||
| |
| − | |-
| |
| − | |||Zirrhose||||
| |
| − | |-
| |
| − | |||Zuschnitt Lamellen Hauptresektat||||
| |
| − | |-
| |
| − | |||Zuschnitt Lamellen Nachresektat||||
| |
| − | |-
| |
| − | |||Zyklustermin||||d
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 23: Attribut-Wert-Paare (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ==66. Codes aus IHE Anatomy Pathology Report==
| |
| − | | |
| − | | |
| − | ===67. Codes für Specimen Types===
| |
| − | | |
| − | nach IHE (IHE APSR Trial Implementation, March 31, 2011) ist ein Specimen collection procedure generic template das Elterntemplate für jedes organspezifische Template.
| |
| − | Jedes organspezifische Template hat Vokabularbegrenzungen, die für das jeweilige Organ spezifisch sind:
| |
| − | | |
| − | Ein Value Set ist gebunden an ein Prozedur Code-Element, das die verschiednen Prozeduren, die an diesem Organ möglich sind, auflistet.
| |
| − | | |
| − | Ein zweiter Value Set ist gebunden and das Prozedut-Target-Site Code-Element, das die möglichen präzisen Lokalisationen an diesem spezifischen Organ auflistet.
| |
| − | | |
| − | z.B.
| |
| − | {| class="hl7table" | |
| − | !(PathLex)Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |2257||Breast-Specimen-Specimen collection procedure
| |
| − | ||Excision with wire-guided localization
| |
| − | |-
| |
| − | |2256||Breast-Specimen-Specimen collection procedure
| |
| − | ||Excision without wire-guided localization
| |
| − | |-
| |
| − | |662||Breast-Specimen-Specimen collection procedure
| |
| − | ||Total mastectomy (including nipple and skin)
| |
| − | |-
| |
| − | |666||Breast-Specimen-Target site
| |
| − | ||Lower inner quadrant
| |
| − | |-
| |
| − | |663||Breast-Specimen-Target site
| |
| − | ||Upper outer quadrant
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 24: HL7 Table 0487 resp. 0700 (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===68. Codes für Specimen Reject Reason===
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !Code!!Codename!!Bedeutung
| |
| − | |-
| |
| − | |1||||
| |
| − | |-
| |
| − | |2||||
| |
| − | |-
| |
| − | |3||||
| |
| − | |-
| |
| − | |4||||
| |
| − | |-
| |
| − | |5||||
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 25: HL7 Table 0490 (OID 1.2.276.0.76.5.??????)
| |
| − | | |
| − | ===69. LOINC Codes===
| |
| − | | |
| − | {| class="hl7table" | |
| − | !LOINC Code!!LOINC Code Name
| |
| − | |-
| |
| − | |22637-3 ||Path report.final diagnosis
| |
| − | |-
| |
| − | |33746-9 ||Pathologic findings
| |
| − | |-
| |
| − | |22636-5 ||Path report.relevant Hx
| |
| − | |-
| |
| − | |22633-2 ||Path report.site of origin
| |
| − | |-
| |
| − | |22634-0 ||Path report.gross description
| |
| − | |-
| |
| − | |22635-7 ||Path report.microscopic observation
| |
| − | |-
| |
| − | |22638-1 ||Path report.comments
| |
| − | |-
| |
| − | |22639-9 ||Path report.supplemental reports
| |
| − | |-
| |
| − | |}
| |
| − | Tabelle 26: LOINC Codes (OID 2.16.840.1.113883.6.1)
| |
| − | | |
| − | =Anhang A: Diverses=
| |
| − | | |
| − | ==Offene Punkte==
| |
| − | * Wikifizierung abschließen
| |
| − | * Codesysteme vervollständigen
| |
| − | * Elemente und Attribute vollständig auflisten (inkl. Wikifizierung)
| |
| − | * Abgleich mit IHE AP und HL7 AP
| |
| − | * ..
| |
| − | | |
| − | ==Beispieldokument==
| |
| − | | |
| − | ===CDA-Header===
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <?xml version="1.0" encoding="UTF-8"?>
| |
| − | <?xml-stylesheet type="text/xsl" href="vhitg-cda-v3.xsl"?>
| |
| − | <ClinicalDocument xmlns="urn:hl7-org:v3"
| |
| − | xmlns:sciphox="urn::sciphox-org/sciphox"
| |
| − | xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
| |
| − | xsi:schemaLocation="urn:hl7-org:v3 CDA.xsd">
| |
| − | <typeId root="2.16.840.1.113883.1.3" extension="POCD_HD000040"/>
| |
| − | <id extension="60467,36049" root="1.2.276.0.58"/>
| |
| − | <code code="11488-4" codeSystem="2.16.840.1.113883.6.1"
| |
| − | displayName="Consultation note"/>
| |
| − | <title>Pathologisch-anatomische Begutachtung mit kritischer Stellungnahme </title>
| |
| − | <effectiveTime value="20100224"/>
| |
| − | <confidentialityCode code="N" codeSystem="2.16.840.1.113883.5.25"/>
| |
| − | <languageCode code="de"/>
| |
| − | <setId extension="D1" root="2.16.840.1.113883.3.933"/>
| |
| − | <versionNumber value="1"/>
| |
| − | <recordTarget>
| |
| − | <!--- Patienten-Daten -->
| |
| − | <patientRole>
| |
| − | <id extension="6" root="2.16.840.1.113883.3.933"/>
| |
| − | <addr>
| |
| − | <streetName>Steinstr.</streetName>
| |
| − | <houseNumber>12</houseNumber>
| |
| − | <postalCode>30156</postalCode>
| |
| − | <city>Hamburg</city>
| |
| − | </addr>
| |
| − | <telecom use="WP" value="tel:040-555-12345"/>
| |
| − | <telecom use="HP" value="tel:040-222-76543"/>
| |
| − | <patient>
| |
| − | <name>
| |
| − | <prefix>Dr.</prefix>
| |
| − | <given>Alfred</given>
| |
| − | <family>Hafer</family>
| |
| − | </name>
| |
| − | <administrativeGenderCode code="M" codeSystem="2.16.840.1.113883.5.1"/>
| |
| − | <birthTime value="19450601"/>
| |
| − | <birthplace>
| |
| − | <place>
| |
| − | <addr>
| |
| − | <city>Sassnitz</city>
| |
| − | </addr>
| |
| − | </place>
| |
| − | </birthplace> </patient>
| |
| − | <providerOrganization>
| |
| − | <telecom use="WP" value="tel:06151-1111111"/>
| |
| − | <telecom use="WP" value="fax:06151-2222222"/>
| |
| − | <addr>
| |
| − | <streetName>Musterstr.</streetName>
| |
| − | <houseNumber>1</houseNumber>
| |
| − | <postalCode>64283</postalCode>
| |
| − | <city>Darmstadt</city>
| |
| − | </addr>
| |
| − | </providerOrganization>
| |
| − | </patientRole>
| |
| − | </recordTarget>
| |
| − | | |
| − | <author>
| |
| − | <!--- author -->
| |
| − | <time value="20060924"/>
| |
| − | <assignedAuthor>
| |
| − | <id extension="6319123" root="2.16.840.1.113883.3.933"/>
| |
| − | <assignedPerson>
| |
| − | <name>
| |
| − | <prefix qualifier="AC">Dr.med.</prefix>
| |
| − | <given>Hans</given>
| |
| − | <family>Topp-Gluecklich</family>
| |
| − | </name>
| |
| − | </assignedPerson>
| |
| − | <representedOrganization>
| |
| − | <name>Praxis Dr.med. Gluecklich </name>
| |
| − | <telecom use="WP" value="tel:06151-1111111"/>
| |
| − | <telecom use="WP" value="fax:06151-2222222"/>
| |
| − | <addr>
| |
| − | <streetName>Musterstr.</streetName>
| |
| − | <houseNumber>1</houseNumber>
| |
| − | <postalCode>64283</postalCode>
| |
| − | <city>Darmstadt</city>
| |
| − | </addr>
| |
| − | </representedOrganization>
| |
| − | </assignedAuthor>
| |
| − | </author>
| |
| − | | |
| − | <custodian>
| |
| − | <!--- Organisation von der das Dokument stammt -->
| |
| − | <assignedCustodian>
| |
| − | <representedCustodianOrganization>
| |
| − | <id extension="M345" root="1.2.276.0.58"/>
| |
| − | <name>Praxis Dr.med. Gluecklich </name>
| |
| − | <telecom nullFlavor="UNK"/>
| |
| − | <addr>
| |
| − | <streetName>Musterstr.</streetName>
| |
| − | <houseNumber>1</houseNumber>
| |
| − | <postalCode>64283</postalCode>
| |
| − | <city>Darmstadt</city>
| |
| − | </addr>
| |
| − | </representedCustodianOrganization>
| |
| − | </assignedCustodian>
| |
| − | </custodian>
| |
| − | | |
| − | <informationRecipient typeCode="PRCP">
| |
| − | <!--- Empfaenger -->
| |
| − | <intendedRecipient classCode="PUB">
| |
| − | <id extension="21233445" root="2.16.840.1.113883.3.933"/>
| |
| − | <receivedOrganization>
| |
| − | <name>Institut für Onkologie des Krankenhauses XYZ</name>
| |
| − | <telecom use="WP" value="fax:02431/901-6210"/>
| |
| − | <addr>
| |
| − | <streetName>Postbox 11 52</streetName>
| |
| − | <postalCode>24099</postalCode>
| |
| − | <city>Kiel</city>
| |
| − | </addr>
| |
| − | </receivedOrganization>
| |
| − | </intendedRecipient>
| |
| − | </informationRecipient>
| |
| − | | |
| − | <legalAuthenticator>
| |
| − | <!--- legalAuthenticator -->
| |
| − | <time value="20060721"/>
| |
| − | <signatureCode code="S"/>
| |
| − | <assignedEntity>
| |
| − | <id extension="6319123" root="2.16.840.1.113883.3.933"/>
| |
| − | <assignedPerson>
| |
| − | <name>
| |
| − | <prefix qualifier="AC">Dr.med.</prefix>
| |
| − | <given>Hans</given>
| |
| − | <family>Topp-Gluecklich</family>
| |
| − | </name>
| |
| − | </assignedPerson>
| |
| − | <representedOrganization>
| |
| − | <name>Praxis Dr.med. Gluecklich </name>
| |
| − | <telecom use="WP" value="fax:061512222222"/>
| |
| − | <addr>
| |
| − | <streetName>Musterstr.</streetName>
| |
| − | <houseNumber>1</houseNumber>
| |
| − | <postalCode>64283</postalCode>
| |
| − | <city>Darmstadt</city>
| |
| − | </addr>
| |
| − | </representedOrganization>
| |
| − | </assignedEntity>
| |
| − | </legalAuthenticator>
| |
| − | | |
| − | <component>
| |
| − | <structuredBody>
| |
| − | ... (s.u.)
| |
| − | </structuredBody>
| |
| − | </component>
| |
| − | </ClinicalDocument>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ===74. CDA-Body===
| |
| − | | |
| − | <syntaxhighlight lang="xml">
| |
| − | <component>
| |
| − | <structuredBody>
| |
| − | <component> <!-- Anrede -->
| |
| − | <section>
| |
| − | <code code="X-SALUT" codeSystem="2.16.840.1.113883.6.1" />
| |
| − | <text>
| |
| − | <paragraph>Sehr geehrter Herr Kollege Dr. Heitmann,</paragraph>
| |
| − | <paragraph>Vielen Dank für die freundliche Überweisung
| |
| − | des Patienten Paul Pappel, geb. 12. Dez. 1955.
| |
| − | </paragraph>
| |
| − | </text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component> <!-- Fragestellung -->
| |
| − | <section>
| |
| − | <code code="X-RFR" codeSystem="2.16.840.1.113883.6.1" />
| |
| − | <title>Fragestellung</title>
| |
| − | <text>Tod verursacht durch onkologische Erkrankung?</text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component>
| |
| − | <!-- Diagnose mit ICD Komponente -->
| |
| − | <section>
| |
| − | <code code="27754-1" codeSystem="2.16.840.1.113883.6.1"
| |
| − | codeSystemName="LOINC"/>
| |
| − | <title>22.02.2010: Diagnosen mit ICD 10</title>
| |
| − | <text>
| |
| − | <paragraph> Diagnose: <content ID="diag-1">????????</content>
| |
| − | </paragraph>
| |
| − | </text>
| |
| − | <entry>
| |
| − | <observation classCode="OBS" moodCode="EVN">
| |
| − | <code code="DISDX" codeSystem="2.16.840.1.113883.3.7.1.16"
| |
| − | codeSystemName="LOINC" displayName="Entlassdiagnosen"/>
| |
| − | <statusCode code="completed"/>
| |
| − | <effectiveTime>
| |
| − | <low value="20050829"/>
| |
| − | </effectiveTime>
| |
| − | <value xsi:type="CD" code="O????"
| |
| − | codeSystem="1.2.276.0.76.5.?????"
| |
| − | codeSystemName="ICD10gm2010"
| |
| − | displayName="?????">
| |
| − | <originalText>
| |
| − | <reference value="\#diag-1"/>
| |
| − | </originalText>
| |
| − | <qualifier>
| |
| − | <name code="8" codeSystem="2.16.840.1.113883.3.7.1"
| |
| − | displayName="Diagnosesicherheit"/>
| |
| − | <value code="G" codeSystem="2.16.840.1.113883.3.7.1.8"
| |
| − | displayName="Gesichert"/>
| |
| − | </qualifier>
| |
| − | </value>
| |
| − | </observation>
| |
| − | </entry>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component>
| |
| − | <section>
| |
| − | <!--
| |
| − | <templateID root="1.2.276.0.76.3.1.81.1.4.xxx"/>
| |
| − | -->
| |
| − | <code code="??????????" codeSystem="1.2.276.0.76.3.1.81.1.3.4.xxxx
| |
| − | codeSystemName="xxxx"/>
| |
| − | <title>xxxx</title>
| |
| − | <text>xxxxxx</text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component>
| |
| − | <!-- Empfehlung -->
| |
| − | <section>
| |
| − | <code code="????????" codeSystem="2.16.840.1.113883.6.1"
| |
| − | codeSystemName="LOINC"/>
| |
| − | <title>Weitergabe</title>
| |
| − | <text>Geben Sie diesen Befund an den behandelnden Arzt weiter.</text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | <component> <!-- Schlusstext -->
| |
| − | <section>
| |
| − | <text> Mit freundlichen, kollegialen Grüßen </text>
| |
| − | </section>
| |
| − | </component>
| |
| − | | |
| − | | |
| − | </structuredBody>
| |
| − | </component>
| |
| − | </syntaxhighlight >
| |
| − | | |
| − | ==75. Referenzen/Literatur==
| |
| − | | |
| − | {| | |
| − | |\[DIMDI, Alpha_Id\]||Alpha-ID - Die Identifikationsnummer, [http://www.dimdi.de/static/de/ehealth/alpha-id/index.htm http://www.dimdi.de/static/de/ehealth/alpha-id/index.htm]
| |
| − | |-
| |
| − | |\[DIMDI, Verschl\]||Anleitung zur Verschlüsselung, [http://www.dimdi.de/static/de/klassi/diagnosen/icd10/icdsgbv20.htm http://www.dimdi.de/static/de/klassi/diagnosen/icd10/icdsgbv20.htm]
| |
| − | |-
| |
| − | |\[DIMDI, Basis\]||Basiswissen Codieren, DIMDI 2004
| |
| − | |-
| |
| − | |\[BMGS, 2004\]||ICD-10-Bekanntmachung des BMGS, [http://www.zi-berlin.de/Zi_ICD10Browser/zi_icd_10_browser.htm http://www.zi-berlin.de/Zi_ICD10Browser/zi_icd_10_browser.htm]
| |
| − | |-
| |
| − | |\[InEK, Codierrichtlinien\]||Deutsche Codierrichtlinien – Version 2005, Institut für Entgeltsystem im Krankenhaus (InEK gGmbH) 2004, [http://www.g-drg.de/service/download/veroeff_2005/DKR2005_Endversion_PDF30_040916_1500.pdf http://www.g-drg.de/service/download/veroeff_2005/DKR2005_Endversion_PDF30_040916_1500.pdf]
| |
| − | |-
| |
| − | |\[HL7 Datentypen\]||HL7 Version 3 Datentypen und CMETs für das Deutsche Gesundheitswesen, www.hl7.de (Publikationen)
| |
| − | |-
| |
| − | |\[CDAr2Arztbrief\]||Arztbrief auf Basis der HL7 Clinical Document Architecture Release 2 für das deutsche Gesundheitswesen, Version 1.50 vom 12.05.2006, herausgegeben vom VHitG, HL7 Deutschland und der Arbeitsgemeinschaft Sciphox, www.hl7.de (Publikationen)
| |
| − | [http://www.hl7.de/download/documents/cdar2-arztbrief/Leitfaden-VHitG-Arztbrief-v150.pdf http://www.hl7.de/download/documents/cdar2-arztbrief/Leitfaden-VHitG-Arztbrief-v150.pdf]
| |
| − | |-
| |
| − | |\[Wiley\]||TNM-System: Wiley Interscience
| |
| − | |-
| |
| − | |}
| |
| − | | |
| − | ==76. Zeitangaben==
| |
| − | In einem Bericht tauchen mehrere Zeitangaben auf, die hier einmal in Form einer Übersicht dargestellt werden sollen. Die Angaben in der Bedingung beziehen sich auf die Nummern aus der ersten Spalte, d.h. hierüber wird eine Reihenfolge etabliert:
| |
| − | | |
| − | {| class="hl7table"
| |
| − | !\#!!Datum!!Art!!Bedingung!!Krankenhaus!!Labor!!HL7 V3<br>(CDA später)
| |
| − | |-
| |
| − | |1||Auftragserfassung||Beginn||||x||||Order
| |
| − | |-
| |
| − | |2||Auftragserfassung||Ende||1 < 2||x||||
| |
| − | |-
| |
| − | |3||Auftragsfreigabe||TS||2 < 3||x||||
| |
| − | |-
| |
| − | |4||Auftragsübermittlung||TS||3 < 4||x||||
| |
| − | |-
| |
| − | |5||Probenentnahme||von/bis||||x||||Specimen Specimen¬Collection¬Process
| |
| − | |-
| |
| − | |6||Probenversand (Ausgang)||TS||5 < 6||x||||
| |
| − | |-
| |
| − | |7||Auftragseingang||TS||4 < 7||||x||Acknow¬ledgement
| |
| − | |-
| |
| − | |8||Auftragsbestätigung||TS||7 < 8||||x||Promise
| |
| − | |-
| |
| − | |9||Probeneingang||TS||6 < 9||||x||Specimen-> Specimen¬Process¬Step
| |
| − | |-
| |
| − | |10||Probenuntersuchung||Beginn||9 < 10 <br> 7 < 10||||x||documentationOf ServiceEvent->
| |
| − | Specimen
| |
| − | |-
| |
| − | |11||Probenuntersuchung||Ende||10 < 11||||x||
| |
| − | |-
| |
| − | |12||Befundung||Beginn||10 < 12||||x||Specimen-> ObservationEvent
| |
| − | author.time
| |
| − | |-
| |
| − | |13||Befundung||Ende||12 < 13||||x||
| |
| − | |-
| |
| − | |14||Niederschrift Befund||Beginn||13 < 14||||x||dataEnterer.time
| |
| − | |-
| |
| − | |15||Niederschrift Befund||Ende||14 < 15||||x||
| |
| − | |-
| |
| − | |16||Freigabe Befund||TS||15 < 16||||x||legalAuthenticator .time
| |
| − | |-
| |
| − | |17||Übermittlung Befund||TS||16 < 17||||x||Wrapper
| |
| − | |-
| |
| − | |18||Befundeingang||TS||17 < 18||x||||Wrapper
| |
| − | |-
| |
| − | |19||Befund gelesen||TS||18 < 19||x||||-
| |
| − | |-
| |
| − | |}
| |
| − | | |
| − | Freigaben können mehrstufig erfolgen.
| |
| − | | |
| − | Mehrere Berichte in Abhängigkeit des Prozessschrittes
| |
| − | | |
| − | =Anhang B: Verzeichnisse=
| |
| − | | |
| − | ==Abbildungverzeichnis==
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ==Tabellenverzeichnis==
| |
| − | | |
| − | tbd
| |
| − | | |
| − | ==Index==
| |
| | | | |
| − | tbd
| + | =References= |
| | + | <references/> |
Dieses Material ist Teil des Leitfadens Implementierungsleitfaden.
- Direkt im Wiki geändert werden sollten Schreibfehler, ergänzende Hinweise.
- Offene Fragen, die der Diskussionen bedürfen, sollten auf der Diskussionsseite aufgenommen werden.
- Liste der Seiten dieses Leitfadens: hier, Liste der Seiten, in denen dieses Material verwendet (transkludiert) siehe hier .
|
Foreword
This is a supplement to the IHE Pathology
and Laboratory Medicine Technical Framework V8.0. Each supplement undergoes a
process of public comment and trial implementation before being incorporated
into the volumes of the Technical Frameworks.
This supplement was published on September 27, 2017 for Public Comment, with a comment period extending to mid-November. The current content is the Trial Implementation version, which has taken in consideration the comments received.
This supplement describes changes to the
existing technical framework documents.
“Boxed” instructions like the sample
below indicate to the Volume Editor how to integrate the relevant section(s)
into the relevant Technical Framework volume.
Replace Section X.X by the following:
Where the amendment adds text, make the added
text bold underline. Where the amendment removes
text, make the removed text bold strikethrough.
When entire new sections are added, introduce with editor’s instructions to
“add new text” or similar, which for readability are not bolded or underlined.
General information about IHE can be found at: http://www.ihe.net.
Information about the IHE Pathology and Laboratory Medicine domain can be found at: http://ihe.net/IHE_Domains.
Information about the structure of IHE Technical Frameworks and Supplements can be found at: http://ihe.net/IHE_Process and http://ihe.net/Profiles.
The current version of the IHE Technical Framework (if applicable) can be found at: http://ihe.net/Technical_Frameworks.
Comments may be submitted on IHE Technical Framework templates any time at http://ihe.net/ihetemplates.cfm. Please enter comments/issues as soon as they are found. Do not wait until a future review cycle is announced.
Acknowledgements
Following authors mainly contributed to this document:
- Francois Macary, PHAST, Paris
- Dr. Gunter Haroske, Federal Association of German Pathologists, Berlin
- Dr. Frank Oemig, Deutsche Telekom Healthcare Solutions GmbH, Bonn
- Dr. Riki Merrick, APHL, San Francisco
- Dr. Raj Dash, Duke University, Durham
They have to acknowledge the contributions of Dr. Kai U. Heitmann, who managed the cooperation between PaLM and ART-DECOR. It was the very first time that a complete IHE TF document could be developed by means of the ART-DECOR tools and a media wiki.
Introduction to this Supplement
This supplement complements volume 1 of
the PaLM technical framework with the description of the APSR 2.0 content
profile, and volume 3 with the bindings, content modules and value sets, which specify
this profile.
Open Issues and Questions
None yet
Closed Issues
APSR-07 – Representing the hierarchy of specimens in an entry : This APSR
supplement enables to represent the hierarchy of specimens at the CDA level 3.
The operations on specimen and production of child specimens are tracked in the
“Procedure Steps” section, which has a level 3 entry.
APSR-10 – Observation related to multiple specimens: For example tumor staging may require combining data from multiple specimens (e.g., a breast excision with positive margins followed by a re-excision with clear margins – in this case the tumor size may be a composite of measurements from both specimens. Another example is staging of ovarian carcinomas with multiple biopsies of pelvis, peritoneum, nodes, omentum, etc.). To accommodate these use
cases, each problem organizer as well as each observation may reference any number of specimens using the <specimen> child element. Each of these references may point to a detailed description of the specimen, in the "procedure steps" section.
APSR-11 – Derivative specimens:Specimens
derived from primary specimens for ancillary studies, which may be sent to a
reference lab or done in another part of the same institution, are included in
the scope of this profile. The results produced on a derived specimen are
attached to this specimen in the report.
VOLUME 1 - PROFILES
10 Anatomic Pathology Structured Report (APSR) Profile
This content profile describes an
anatomic pathology structured report (APSR) as a digital document to be shared or
exchanged between pathology laboratories and other care providers and
institutions.
Anatomic pathology structured reports
document the findings on specimens removed from patients for diagnostic or
therapeutic reasons. This information can be used for patient care, clinical
research and epidemiology. Standardizing and computerizing anatomic pathology
reports is necessary to improve the quality of reporting and to facilitate the
exchange and reuse of the content of these reports.
This content profile describes a digital
anatomic pathology report shared in a human-readable format, which may include
images, and which also contains findings and observations in a machine-readable
format, to facilitate the integration of these into the database of a consumer
system, and to enable the application of automated reasoning to this content.
The scope of this IHE content profile covers all fields
of anatomic pathology (cancers, benign neoplasms as well as non-neoplastic
conditions) as well as cytopathology.
Goldsmith J.D. et al.[1] is the first source of specification for
this content profile. This article delineates the required, preferred, and
optional elements which should be included in any report of surgical pathology.
This source is complemented by the
“Cancer Checklists” produced by the College of American Pathologists[2], and by
the “comptes rendus d’anatomopathologie : données minimales à renseigner pour
une tumeur primitive” produced by the French society of pathology (SFP[3])
for the French cancer institute (INCa [4]), and by the German "Guideline Pathology / Neuropathology" (formerly TM-30) of the Sector Committee Pathology for the implementation of DIN EN ISO/EC 17020.
This profile has also benefited from the
guidance on cancer AP reports provided by the North-American Association of
Central Cancer Registries; some of the example snippets captured in the profile
leverage the NAACCR Standards for Cancer Registries, Volume V, Pathology
Laboratory Electronic Reporting.
10.1 APSR Actors/Transactions
This
section defines the actors, transactions, and/or content modules in this
profile. General definitions of actors are given in the Technical Frameworks
General Introduction Appendix A published here.
Figure 10.1-1
shows the actors directly involved in the APSR Profile and the direction that
the content is exchanged.
A
product implementation using this profile must group actors from this profile
with actors from a workflow or transport profile to be functional. The grouping
of the content module described in this profile to specific actors is described
in more detail in the “Required Actor Groupings” section below.
Figure 10.1-1 APSR Actor Diagram
Table 10.1-1 lists the content module(s)
defined in the APSR profile. To claim support with this profile, an actor shall
support all required content modules (labeled “R”) and may support optional
content modules (labeled “O”).
Table 10.1-1: <Profile Acronym>Profile - Actors and Content Modules
| Actors |
Content Modules |
Optionality |
Reference
|
| Content
Creator
|
Anatomic Pathology Structured Report
1.3.6.1.4.1.19376.1.8.1.1.1
|
R
|
PaLM TF-3: 6.3.1.2
|
| Content
Consumer
|
Anatomic Pathology Structured Report
1.3.6.1.4.1.19376.1.8.1.1.1
|
R
|
PaLM TF-3: 6.3.1.2
|
10.1.1 Actor Descriptions and Actor Profile Requirements
Most requirements are documented in
Content Modules (Volume 3). This section documents any additional requirements
on profile’s actors.
10.2 APSR Actor Options
Options that may be selected for each
actor in this profile are listed in the table 10.2-1. These options are further
detailed in PCC Technical Framework Volume 2 as indicated in the rightmost
column.
Table 10.2-1 Anatomic Pathology Structured Report - Actors and Options
| Actor |
Option Name |
Reference
|
| Content
Creator
|
None
|
|
| Content
Consumer
|
View Option (1)
Document Import Option (1)
Section Import Option (1)
|
PCC TF-2:3.1.1
PCC TF-2:3.1.2
PCC TF-2:3.1.3
|
Note 1: The Content Consumer Actor shall support at
least one of these options.
10.3 APSR Required Actor Groupings
An Actor from this profile (Column 1)
shall implement all of the required transactions and/or content modules in this
profile in addition to all of the transactions required for the grouped
actor (Column 2).
In some cases, required groupings are
defined as at least one of an enumerated set of possible actors; this is
designated by merging column one into a single cell spanning multiple potential
grouped actors. Notes are used to highlight this situation.
Section 10.5 describes some optional
groupings that may be of interest for security considerations and section 10.6
describes some optional groupings in other related profiles.
Table 10.3-1: Anatomic Pathology Structured Report - Required Actor Groupings
| APSR Actor |
Actor to be grouped with |
Reference |
Content Bindings Reference
|
| Content
Creator
|
ITI XDS.b
Document Source
OR
ITI XDM Portable Media Creator
OR
ITI XDR Document Source
OR
ITI MHD Document Source
|
ITI TF-1:10
ITI TF-1:16
ITI TF-1:15
ITI TF-1:33
|
|
| Content
Consumer
|
ITI XDS.b Document Consumer
OR
ITI XDM Portable Media Consumer
OR
ITI XDR Document
Recipient
OR
ITI MHD Document Consumer
|
ITI TF-1:10
ITI TF-1:16
ITI TF-1:15
ITI TF-1:33
|
|
Note 1: Each
actor of APSR SHALL be grouped with at least one of the ITI actors listed in
its table row.
10.4 APSR Overview
10.4.1 Concepts
This content profile represents a common
digital document model applicable to any structured report for surgical
pathology in all fields of anatomic pathology (cancers, benign neoplasms, non-neoplastic
conditions) as well as for cytopathology.
This common model is composed of a header conveying
the context of care (patient, care providers, pathologists, laboratories,
order, act documented …) and a body. The body organizes the human-readable
content of the report in a number of sections. Each section may also provide
machine-readable content in a repeatable “entry” embedded in the section. This common model defines the order of
appearance, cardinalities and internal structure of each section, and of each
entry embedded in each section.
Figure 10.4.1-1 shows this general model
applicable to any pathology digital report.
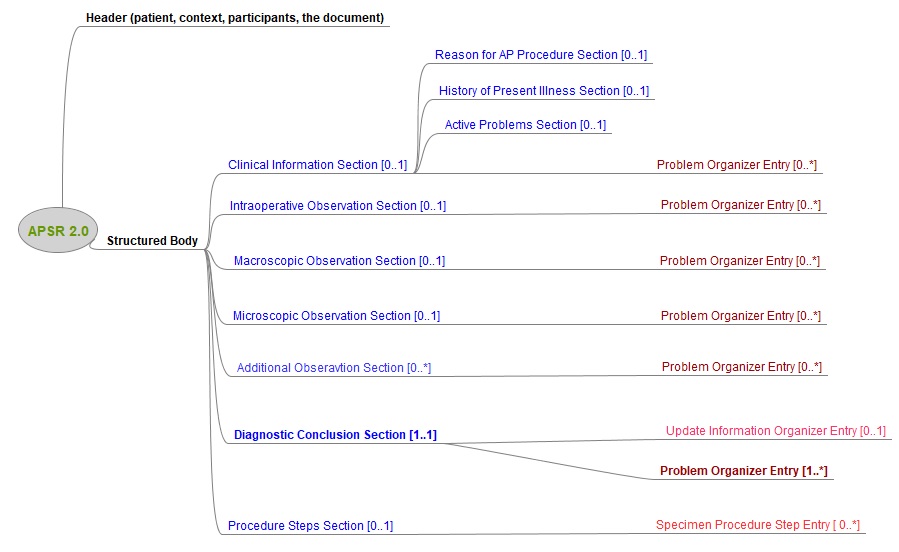
Figure 10.4.1-1
Common model for a digital anatomic pathology structured report
Note 1: The only section that is mandatory is
the Diagnostic Conclusion section.
10.4.2 Use Cases
10.4.2.1 Use case #1: Single Report
Anatomic pathology order fulfilled by a pathology laboratory produces a report.
10.4.2.1.1 Single Report Use Case Description
Dr. Eva Surgeon, PhD, takes a ultrasound guided core biopsy from a breast tumor from patient Eve Onewoman, born on Sept 21 1971, requests the procedure “breast core biopsy specimen - pathological examination” and sends the specimen to the anatomic pathology laboratory of the Cancer Institute.
One specimen (five cores) is accessioned by the anatomic pathology laboratory under the accession number A710240008. The staff performs a macroscopic examination of the specimen; gross imaging is performed if needed. The specimen with the specimen ID A710240008_A is processed for microscopic examination and other special ancillary techniques or tissue banking if needed. During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process of the macroscopic and microscopic observations and some ancillary techniques, done by the pathologists Dr. Marcel Pathologist, PhD, and Dr. Jonas Jones, M.D., Dr. Marcel Pathologist queries the Content Creator application for the appropriate APSR template, fills the form, binds some relevant images and/or regions of interest to specific observations, validates and signs the digital report.
10.4.2.1.2 Single Report Process Flow
Intentionally left blank
10.4.2.1.3 Single Report Text Example
Macroscopic observation
A. "RIGHT BREAST FIVE CORES 8-9:00" (ULTRASOUND GUIDED NEEDLE CORE BIOPSY):
Microscopic observation
INVASIVE ADENOCARCINOMA OF THE BREAST.
ICD-O-3-CLASSIFICATION: C50.3 M8500/33
HISTOLOGIC TYPE: DUCTAL.
NOTTINGHAM COMBINED HISTOLOGIC GRADE: 1 OF 3.
TUBULE FORMATION SCORE: 2.
NUCLEAR PLEOMORPHISM SCORE: 2.
MITOTIC RATE SCORE: 1.
IN-SITU CARCINOMA: EQUIVOCAL.
BREAST CANCER BIOMARKER STUDIES:
PARAFFIN BLOCK NUMBER: A1.
ER INTERPRETATION: POSITIVE ESTROGEN RECEPTOR ACTIVITY (ALLRED SCORE = 8, Percentage of positive cells = 85%, Staining Intensity score = 3).
PR INTERPRETATION: POSITIVE PROGESTERONE RECEPTOR ACTIVITY (ALLRED SCORE = 8).
DAKO EGFR PHARMDX IMMUNOHISTOCHEMISTRY: NEGATIVE (0) FOR EXPRESSION OF
EPIDERMAL GROWTH FACTOR RECEPTOR.
HER2/NEU IMMUNOHISTOCHEMISTRY: AMBIGUOUS(2+) FOR OVEREXPRESSION OF HER2/NEU
ONCOPROTEIN.
HER2/NEU FISH RESULT: NEGATIVE FOR AMPLIFICATION OF HER2/NEU.
Diagnostic conclusion
A. "RIGHT BREAST FIVE CORES 8-9:00" (ULTRASOUND GUIDED NEEDLE CORE BIOPSY):
INVASIVE ADENOCARCINOMA OF THE BREAST.
ICD-O-3-CLASSIFICATION: C50.3 M8500/33
HISTOLOGIC TYPE: DUCTAL.
NOTTINGHAM COMBINED HISTOLOGIC GRADE: 1 OF 3.
TUBULE FORMATION SCORE: 2.
NUCLEAR PLEOMORPHISM SCORE: 2.
MITOTIC RATE SCORE: 1.
IN-SITU CARCINOMA: EQUIVOCAL.
BREAST CANCER BIOMARKER STUDIES:
PARAFFIN BLOCK NUMBER: A1.
ER INTERPRETATION: POSITIVE ESTROGEN RECEPTOR ACTIVITY (ALLRED SCORE = 8, Percentage of positive cells = 85%, Staining Intensity score = 3).
PR INTERPRETATION: POSITIVE PROGESTERONE RECEPTOR ACTIVITY (ALLRED SCORE = 8).
DAKO EGFR PHARMDX IMMUNOHISTOCHEMISTRY: NEGATIVE (0) FOR EXPRESSION OF
EPIDERMAL GROWTH FACTOR RECEPTOR.
HER2/NEU IMMUNOHISTOCHEMISTRY: AMBIGUOUS(2+) FOR OVEREXPRESSION OF HER2/NEU
ONCOPROTEIN.
HER2/NEU FISH RESULT: NEGATIVE FOR AMPLIFICATION OF HER2/NEU.
Procedure steps:
RIGHT BREAST FIVE CORES 8-9:00" (ULTRASOUND GUIDED NEEDLE CORE BIOPSY) PARAFFIN BLOCK NUMBER: A1.:FOUR SECTIONS FOR EACH STAIN:
RIGHT BREAST FIVE CORES 8-9:00
PARAFFIN BLOCK NUMBER: A1
slide from block A1 HE stained
slide from block A1 ER Immunohistochemistry
slide from block A1 PR Immunohistochemistry
slide from block A1 EGFR (PharmDX) Immunohistochemistry
slide from block A1 HER2 Immunohistochemistry
slide block from A1 HER2 FISH
10.4.2.2 Use Case #2: Multi-step Report
Reporting includes multiple successive steps.
10.4.2.2.1 Multi-step Report Use Case Description
A surgeon removes a breast tumor from a patient, requests the procedure “breast surgical specimen - frozen sections & pathological examination”, and “breast surgical specimen - pathological examination” and sends the specimen(s) to the anatomic pathology laboratory.
Specimens are accessioned by the anatomic pathology department. The staff performs a macroscopic examination of the specimens, gross imaging is performed if needed. The specimens are processed for intraoperative observation if needed, and tissue banking if needed (e.g., for research purpose). During the imaging interpretation process of frozen sections, microscopic imaging is performed if needed. At the end of the interpretation process, the pathologist queries the Content Creator for the appropriate APSR template, fills the intraoperative observation section, binds some relevant images and/or regions of interest to specific observation(s) if needed, validates and signs (i.e., legally authenticates) the preliminary APSR.
The day after, the specimen(s) are processed for microscopic examination and other special ancillary techniques if needed. During the imaging interpretation process, microscopic imaging is performed if needed. At the end of the interpretation process, pathologist queries the Content Creator for the preliminary APSR, fills the form, binds some relevant images and/or regions of interest to specific observation(s), validates and signs (i.e., legally authenticates) the final APSR.
10.4.2.2.2 Multi-step Report Process Flow
Intentionally left blank
10.5 APSR Security Considerations
See Appendix A of PaLM TF-1.
10.6 APSR Cross Profile Considerations
Intentionally left blank
VOLUME 3 – CONTENT MODULES
1 Introduction
The content of this entire section is identical to the actual section 1 of Volume 3 of PaLM TF.
1.1 Overview of the Anatomic Pathology Technical Framework
Integrating the Healthcare Enterprise (IHE) is an international initiative to promote the use of standards to achieve interoperability among health information technology (HIT) systems and effective use of electronic health records (EHRs). IHE provides a forum for care providers, HIT experts and other stakeholders in several clinical and operational domains to reach consensus on standards-based solutions to critical interoperability issues.
The primary output of IHE is system implementation guides, called IHE Profiles. IHE publishes each profile through a well-defined process of public review and trial implementation and gathers profiles that have reached final text status into an IHE Technical Framework, of which this volume is a part.
For more general information regarding IHE, refer to IHE International website. It is strongly recommended that, prior to reading this volume, the reader familiarizes themselves with the concepts defined in the IHE Technical Frameworks General Introduction.
1.2 Intended Audience
The intended audience of IHE Technical Frameworks Volume 3 is:
- IT departments of healthcare institutions
- Technical staff of vendors participating in the IHE initiative
- Experts involved in standards development
1.3 Overview of Technical Framework Volume 3
Volume 3 is comprised of several distinct sections:
- Section 1 provides background and reference material.
- Section 2 presents the conventions used in this volume to define the content modules.
- Section 3 provides an overview of Content Modules and the terminology used.
- Section 4 is reserved for domain unique Content Module specifications.
- Section 5 lists the namespaces and identifiers defined or referenced and the vocabularies defined or referenced herein.
- Section 6 defines the PaLM domain’s HL7 V3 CDA Content Modules in detail.
- Section 7 defines the PaLM domain’s DICOM content modules in detail.
- Section 8 defines other types of content modules.
The appendices in Volume 3 provide clarification of technical details of the IHE data model and transactions. A glossary of terms and acronyms used in the IHE Technical Framework, including those from relevant standards, is provided in the IHE Technical Frameworks General Introduction. Due to the length of the document, some domains may divide Volume 3 into smaller volumes labeled 3a, 3b, etc. In this case, the Volume 3 appendices are gathered in Volume 3x. Code and message samples may also be stored on the IHE ftp server. In this case, explicit links to the ftp server will be provided in the transaction text.
IHE International welcomes comments on this document and the IHE initiative. They can be submitted by sending an email to the co-chairs and secretary of the Pathology and Laboratory Medicine domain committees at palm@ihe.net
1.5 Copyright Licenses
IHE International hereby grants to each Member Organization, and to any other user of these documents, an irrevocable, worldwide, perpetual, royalty-free, nontransferable, nonexclusive, non-sublicensable license under its copyrights in any IHE profiles and Technical Framework documents, as well as any additional copyrighted materials that will be owned by IHE International and will be made available for use by Member Organizations, to reproduce and distribute (in any and all print, electronic or other means of reproduction, storage or transmission) such IHE Technical Documents.
The licenses covered by this Copyright License are only to those copyrights owned or controlled by IHE International itself. If parts of the Technical Framework are included in products that also include materials owned or controlled by other parties, licenses to use those products are beyond the scope of this IHE document and would have to be obtained from that other party.
1.5.1 Copyright of Base Standards
IHE technical documents refer to and make use of a number of standards developed and published by several standards development organizations. All rights for their respective base standards are reserved by these organizations. This agreement does not supersede any copyright provisions applicable to such base standards.
Health Level Seven, Inc. has granted permission to IHE to reproduce tables from the HL7 standard. The HL7 tables in this document are copyrighted by Health Level Seven, Inc. All rights reserved. Material drawn from these documents is credited where used.
1.6 Trademark
IHE® and the IHE logo are trademarks of the Healthcare Information Management Systems Society in the United States and trademarks of IHE Europe in the European Community. They may only be used with the written consent of the IHE International Board Operations Committee, which may be given to a Member Organization in broad terms for any use that is consistent with the IHE mission and operating principles.
1.7 Disclaimer Regarding Patent Rights
Attention is called to the possibility that implementation of the specifications in this document may require use of subject matter covered by patent rights. By publication of this document, no position is taken with respect to the existence or validity of any patent rights in connection therewith. IHE International is not responsible for identifying Necessary Patent Claims for which a license may be required, for conducting inquiries into the legal validity or scope of Patents Claims or determining whether any licensing terms or conditions provided in connection with submission of a Letter of Assurance, if any, or in any licensing agreements are reasonable or non-discriminatory. Users of the specifications in this document are expressly advised that determination of the validity of any patent rights, and the risk of infringement of such rights, is entirely their own responsibility. Further information about the IHE International patent disclosure process including links to forms for making disclosures is available here. Please address questions about the patent disclosure process to the secretary of the IHE International Board: secretary@ihe.net
1.8 History of Document Changes
Content Modules for the APSR 2.0 Profile:
- Reconfiguration of the Document content module
- Reconfiguration of the Procedure step section content module
- Introduction of the Additional Specified Observation section content module
- Reconfiguration of the entry content modules "Problem organizer", "Specimen procedure steps", and "Update information organizer"
- Introduction of child element content modules "X-specimen identified", "UICC/AJCC-TNM observation", "ICD-O-3 observation", "Assessment scales observation", "Pertinent insurance information".
- Renewal and completion of value sets.
Transformation of a real-world use case into a valid APSR 2.0 xml instance.
Finishing the Supplement ready to trial implementation.
2 Conventions
This document has adopted the following conventions for representing the framework concepts and specifying how the standards upon which the IHE Technical Framework is based shall be applied.
2.1 Content Module Modeling and Profiling Conventions
In order to maintain consistent documentation, modeling methods for IHE content modules and profiling conventions for frequently used standards are maintained as an appendix in the IHE Technical Frameworks General Introduction. Methods described include the standards conventions DICOM, HL7 v2.x, HL7 Clinical Document Architecture (CDA) Documents, etc. These conventions are critical to understanding this volume and should be reviewed prior to reading this text.
2.2 Additional Standards Profiling Conventions
This section defines profiling conventions for standards which are not described in the IHE Technical Frameworks General Introduction.
Not Applicable.
3 Content Modules Overview and Terminology
In the future, an appendix to the IHE Technical Frameworks General Introduction will provide an overview of Content Modules. In the interim, information may be available here.
The Pathology and Laboratory Medicine content modules are listed in the table below:
Table 3-1: Pathology and Laboratory Medicine Content Modules
| Content Module Acronym |
Type of Content Modules |
Semantic |
Status
|
| XD-LAB |
CDA R2 medical document |
Clinical Laboratory Structured Report |
Final Text
|
| APSR |
CDA R2 medical document |
Anatomic Pathology Structured Report |
Trial Implementation
|
4 IHE Pathology and Laboratory Medicine Bindings
4.1 Medical Document Binding to XDS, XDM and XDR
The bindings of the content modules of the PaLM domain leverage the bindings specified by the Patient Care Coordination domain, in PCC TF Volume 2, section 4, with the addition of the constraints specified below.
4.1.1 XDSDocumentEntry Metadata
4.1.1.1 XDSDocumentEntry.eventCodeList
Append this paragraph at the end of the section.
For the APSR content module, The XDSDocumentEntry.eventCodeList provides a means to index anatomic pathology reports by reportable conditions (e.g., certain types of tumors…) so as to facilitate later queries in a registry of shared clinical documents. The conclusions coded in the entry element of the Diagnostic Conclusion section are good candidates for this metadata.
4.1.1.2 XDSDocumentEntry.formatCode
Append this paragraph at the end of the section.
For the APSR content module, The XDSDocumentEntry.formatCode SHALL be urn:ihe:palm:apsr:2016
The associated codingScheme SHALL be 1.3.6.1.4.1.19376.1.2.3
4.1.1.3 XDSDocumentEntry.parentDocumentRelationship
Append this paragraph at the end of the section.
For the APSR content module XDSDocumentEntry.parentDocumentRelationship is constrained to the "RPLC" value. When there is a parent document the current document is a new version of the parent document, replacing it.
- Note 1: A non-final anatomic pathology report published in an XDS infrastructure will likely be replaced afterwards by the final report. When this event occurs, the Content Creator Actor SHALL apply the following rules:
- ClinicalDocument/setId SHALL have the same value in the new report as in the replaced report.
- ClinicalDocument/versionNumber SHALL be incremented in the replacing report (i.e. the final one).
- ClinicalDocument/relatedDocument@typeCode attribute SHALL be valued ”RPLC”
- ClinicalDocument/relatedDocument/parentDocument/id in the new report SHALL be equal to ClinicalDocument/ id of the replaced document.
The Document Source Actor SHALL apply the following rules on XDSDocumentEntry metadata:
- The final report SHALL be associated with the previously published one, using RPLC relationship and the previous report SHALL be “Deprecated” as described in ITI TF-2:4.1.6.1.
- Note 2: A non-final report can also be replaced by a more recent, albeit still non-final report. The rules above also apply in this case.
- Note 3: A final report can also be replaced by a corrective final report. The rules above also apply in this case.
- Note 4: A new version of a report SHOULD have an Update Organizer <entry> in its Diagnostic Conclusion <section> carrying information about what has been changed in comparison with the immediate previous report, and what is the clinical significance of that change.
5 Namespaces and Vocabularies
5.1 OID tree of PAT TF
1.3.6.1.4.1.19376.1.81.3.6.1.4.1.19376.1.8 is the OID of the former IHE Anatomic Pathology domain, whereas 1.3.6.1.4.1.19376.1.81.3.6.1.4.1.19376.1.3 is the OID for PaLM domain :
All exchangeable objects specified by these domains are identified by OIDs built on these roots:
Branch 1.3.6.1.4.1.19376.1.8.1 is dedicated to CDA Content Modules created by the AP domain
Sub-branch 1.3.6.1.4.1.19376.1.8.1.1 is the OID of the generic Document Content Module
Sub-branch 1.3.6.1.4.1.19376.1.8.1.2 is dedicated to Section Content Modules
Sub-branch 1.3.6.1.4.1.19376.1.8.1.3 is dedicated to Entry Content Modules.
Sub-branch 1.3.6.1.4.1.19376.1.8.1.3.6 is the OID of the Problem Organizer
Sub-branch 1.3.6.1.4.1.19376.1.8.1.4 is dedicated to Element Content Modules
Sub-branch 1.3.6.1.4.1.19376.1.8.1.4.9 is the OID of the generic anatomic pathology (AP) observation template
Branch 1.3.6.1.4.1.19376.1.8.2 is dedicated to terminologies defined by AP domain
Sub-branch 1.3.6.1.4.1.19376.1.8.2.1 is dedicated to PathLex, BUT THIS TEMPORARY VOCABULARY IS NO LONGER USED
Branch 1.3.6.1.4.1.19376.1.8.5 is dedicated to Value Sets defined by AP domain.
Branch 1.3.6.1.4.1.19376.1.3.10 is dedicated to Templates newly defined by PaLM domain.
Sub-branch 1.3.6.1.4.1.19376.1.3.10.1 is dedicated to CDA Document Level Templates
Sub-branch 1.3.6.1.4.1.19376.1.3.10.2 is dedicated to CDA Header Level Templates
Sub-branch 1.3.6.1.4.1.19376.1.3.10.3 is dedicated to CDA Section Level Templates
Sub-branch 1.3.6.1.4.1.19376.1.3.10.4 is dedicated to CDA Entry Level Templates
Sub-branch 1.3.6.1.4.1.19376.1.3.10.9 is dedicated to CDA Template Fragments/Supporting Templates
Branch 1.3.6.1.4.1.19376.1.3.11 is dedicated to Value Sets newly defined by PaLM domain.
Branch 1.3.6.1.4.1.19376.1.8.9 is used to identify instances in the examples of AP content built by the PaLM domain.
Notes on other IHE OIDs used in the AP domain:
Branch 1.3.6.1.4.1.19376.1.3.4 is used to identify instances in the examples of AP content built by XD-LAB in the PaLM domain.
5.2 Terminologies and controlled coded vocabularies
This section lists the terminologies and the coded vocabularies referenced by this Volume 3.
Table 5.2-1 Anatomic Pathology Terminologies and Coded Vocabularies
| codeSystem |
codeSystemName and Description |
Owner
|
| 2.16.840.1.113883.6.1
|
LOINC
Logical Observation Identifier Names and Codes
|
Regenstrief Institute
|
| 2.16.840.1.113883.6.8
|
UCUM
Unified Code for Units of Measure
|
Regenstrief Institute, and the UCUM Organization
|
| 2.16.840.1.113883.6.96
|
SNOMED-CT
Systematized Nomenclature of Medicine – Clinical Terms
|
IHTSDO/Snomed International
|
| 1.3.6.1.4.1.19376.1.5.3.2
|
IHEActCode
Vocabulary defined by IHE PCC in PCC TF-2:5.1.2, used for representing annotation comments in the report
|
IHE PCC
|
| 2.16.840.1.113883.5.4
|
Act Code
Codesystem for Acts
|
HL7 International
|
| 2.16.840.1.113883.5.6
|
ActClass
Codesystem for Act Classes
|
HL7 International
|
| 2.16.840.1.113883.5.1052
|
Act Site
Codesystem for Act Sites
|
HL7 International
|
| 2.16.840.1.113883.5.1065
|
ProcedureMethod
Codesystem for Procedure Methods
|
HL7 International
|
| 2.16.840.1.113883.5.111
|
Role Code
Codesystem for Role Codes
|
HL7 International
|
| 2.16.840.1.113883.5.129
|
SpecimenType
Codesystem for Specimen Types
|
HL7
|
| 2.16.840.1.113883.5.83
|
ObservationInterpretation
One or more codes specifying a rough qualitative interpretation of the observation, such as "normal", "abnormal", "below normal", "change up", "resistant", "susceptible", etc.
|
HL7 International
|
| 2.16.840.1.113883.5.84
|
ObservationMethod
A code system that provides additional detail about the means or technique used to ascertain the observation
|
HL7 International
|
| 2.16.840.1.113883.6.3
|
ICD-10
International Classification of Diseases revision 10
|
WHO
|
| 2.16.840.1.113883.6.43.1
|
ICD-O-3
International Classification of Diseases for Oncology, 3rd edition, revision 1, 2013
|
WHO
|
| 1.2.840.10008.2.16.4
|
DICOM controlled vocabulary
The meanings of codes defined in DICOM, either explicitly or by reference to another part of DICOM or an external reference document or standard [5]
|
DICOM
|
| 2.16.840.1.113883.15.16
|
TNM 8th edition
Internationally agreed-upon standards to describe and categorize cancer stages and progression [6]
|
Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC)
|
| 2.16.840.1.113883.15.6
|
TNM 7th edition
Internationally agreed-upon standards to describe and categorize cancer stages and progression [6]
|
Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC)
|
| 2.16.840.1.113883.15.7
|
TNM 6th edition
Internationally agreed-upon standards to describe and categorize cancer stages and progression [6]
|
Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC)
|
| 2.16.840.1.113883.15.8
|
TNM 5th edition
Internationally agreed-upon standards to describe and categorize cancer stages and progression [6]
|
Union for International Cancer Control (UICC) & American Joint Committee on Cancer (AJCC)
|
| 2.16.840.1.113883.6.4
|
ICD-10-PCS
International Classification of Diseases, 10th Revision, Procedure Coding System (ICD-10-PCS)
|
WHO
|
| 1.2.276.0.76.5.464
|
OPS 2017
Code lists to describe and categorize surgeries and procedures, adapted from WHO ICPM for Germany
|
DIMDI / (WHO)
|
| 2.16.840.1.113883.6.174
|
OMIM
Code lists to describe and categorize human genes. OMIM is a comprehensive, authoritative compendium of human genes and genetic phenotypes that is freely available and updated daily.[7]
|
OMIM Johns Hopkins University School of Medicine
|
| 1.2.276.0.76.3.1.131.1.5.1
|
DKG Coding Scheme
Internationally agreed-upon code lists to describe and categorize cancer grading systems, adapted for Germany [8]
|
DKG (Deutsche Krebsgesellschaft)
|
| 1.2.276.0.76.5.336
|
Grading / Differentiation scheme
Code lists to describe and categorize Tumor grading according ICD-O-3, adapted for Germany
|
HL7 Germany
|
| 1.2.276.0.76.5.401
|
Localization scheme for distant metastases
Code lists to describe and categorize localization of metastases according UICC, adapted for Germany
|
HL7 Germany
|
5.3 Value Sets
The value sets defined or referenced by this Volume 3 of the IHE PaLM TF Suppl. are listed and specified in Appendix A of this Volume.
5.4 Namespaces
5.4.1 Namespace protecting extensions to the CDA schema
There is currently one single extension to the CDA.xsd schema used in PaLM TF-3. This extension has been created by former IHE LAB and is protected by this particular namespace in document instances: xmlns:lab="urn:oid:1.3.6.1.4.1.19376.1.3.2"
5.5 References to Content Modules built outside of IHE PAT TF
The Content Modules specified in this Volume 3 of the PAT TF leverage a number of Content Modules (currently CDA templates) produced and maintained by other groups, including other domains of IHE. Table 5.5-1 lists them.
Table 5.5-1 External Content Modules referenced by PAT TF-3
| templateId |
Standard |
Definition |
Source of Specification
|
| 1.3.6.1.4.1.19376.1.5.3.1.3.1 |
CDA R2
|
Reason for referral
|
IHE PCC TF-2:6.3.3.1.2
|
| 1.3.6.1.4.1.19376.1.5.3.1.3.4
|
CDA R2 |
History of present illness
|
IHE PCC TF-2:6.3.3.2.1
|
| 1.3.6.1.4.1.19376.1.5.3.1.3.6
|
CDA R2 |
Active Problems
|
IHE PCC TF-2:6.3.3.2.3
|
| 1.3.6.1.4.1.19376.1.5.3.1.4.2
|
CDA R2 |
Comment
|
IHE PCC TF-2:6.3.4.6
|
| 1.3.6.1.4.1.19376.1.3.3.1.1
|
CDA R2 |
Human Patient
|
IHE PaLM TF-3:6.3.2.11.1
|
| 1.3.6.1.4.1.19376.1.3.1.3
|
CDA R2 |
Specimen Received
|
IHE PaLM TF-3:6.3.4.6
|
| 1.3.6.1.4.1.19376.1.3.1.6
|
CDA R2 |
Laboratory Observation
|
IHE PaLM TF-3:6.3.4.13
|
| 1.3.6.1.4.1.19376.1.3.3.1.4
|
CDA R2 |
Intended Recipient
|
IHE PaLM TF-3:6.3.2.14
|
| 1.3.6.1.4.1.19376.1.3.3.1.6
|
CDA R2 |
Ordering Provider (ordering physician)
|
IHE PaLM TF-3:6.3.2.17
|
| 1.3.6.1.4.1.19376.1.3.3.1.7
|
CDA R2 |
Laboratory Performer
|
IHE PaLM TF-3:6.3.2.20
|
5.6 IHE code and formatCode for Anatomic Pathology Document Template
Merge the content of this section into the current section 5.1.1 of PaLM TF volume 3.
Any AP structured report SHALL be associated with the metadata typeCode = “11526-1”, which is the LOINC code for “Pathology study”.
The table below lists the format codes, template identifiers and media types used by the IHE Profiles specified in the PaLM Technical Framework.
Note that the code system for these codes is 1.3.6.1.4.1.19376.1.2.3 as assigned by the ITI Domain for codes used for the purposes of cross-enterprise document sharing (XDS). For more information see XDS Coding System (1.3.6.1.4.1.19376.1.2.3).
6 PaLM HL7 CDA Content Modules
6.1 Conventions
- In all Content Modules specified in this section, the abbreviation “AP” stands for “Anatomic Pathology”.
6.2 Folder Modules
Intentionally left blank
6.3 Content Modules
This section defines each IHE Pathology and Laboratory Medicine Content Module in detail, specifying the standards used and the information defined.
6.3.1 CDA Document Content Modules
All persons (including the patient) and organizations mentioned in the CDA Document Content Modules SHALL include the elements name, addr and telecom.
6.3.1.1 Clinical Laboratory Report Content Module 1.3.6.1.4.1.19376.1.3.3
Section unchanged, as in PaLM TF-3
6.3.1.2 Anatomic Pathology Structured Report Content Module 1.3.6.1.4.1.19376.1.8.1.1.1
This Content Integration Profile describes a surgical pathology report as an electronic document to be published towards a document sharing resource such as an Electronic Health Record (EHR) or Personal Health Record (PHR) shared by a community of care providers, using one of the document sharing profiles defined in ITI-TF.
Such an electronic document contains the set of releasable results produced by an surgical pathology laboratory in fulfillment of an Order or an Order Group for a patient. The report is shared in a human-readable format. In addition, this electronic anatomic pathology report SHALL contain diagnostic conclusions in a machine-readable format, to facilitate the integration of these observations in the database of a consumer system.
This Document Content Module defines the base set of constraints that apply to all AP structured reports, related to any kind of lesion or diagnostic problem (cancers, benign neoplasms as well as non-neoplastic conditions) as well as for Cytopathology.
In other words, this is the generic template for any AP structured report.
This document content module is identified by templateId 1.3.6.1.4.1.19376.1.8.1.1.1.
The body of this Document Content Module specifies a common hierarchy of sections and entries depicted by figure 10.4.1-1 in Volume 1. The only mandatory section is the Diagnostic Conclusion Section. And the only mandatory entry is the Problem Organizer Entry below this section.
The specification of this Document Content Module is built and published on Art-Decor Pathology Structured Reporting, including schematron rules, code systems and value sets.
| Id | 1.3.6.1.4.1.19376.1.8.1.1.1 | Effective Date | valid from 2014‑05‑13 11:57:57 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | AnatomicPathologyStructuredReportContentModule | Display Name | Anatomic Pathology Structured Report Content Module |
|---|
| Description | Anatomic Pathology Structured Report Content Module.
This document content module represents the generic set of constraints applied to any structured report for surgical pathology in all fields of anatomic pathology (cancers, benign neoplasms as well as non-neoplastic conditions) as well as for Cytopathology.
The body of this Document Content Module specifies a common hierarchy of sections and entries depicted by figure 10.4.1-1 in Volume 1 IHE_PaLM_Suppl_APSR 2.0. The only mandatory section is the Diagnostic Conclusion Section. And the only mandatory entry is the Problem Organizer Entry below this section.
|
|
| Context | Pathname / |
|---|
| Label | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module
|
|---|
| Classification | CDA Document Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 0 templates, Uses 20 templates | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.3.1.1 | Include |  Human Patient (2017) Human Patient (2017) | 2017‑06‑07 | | 1.3.6.1.4.1.19376.1.8.1.4.2 | Include |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 2.16.840.1.113883.10.12.103 | Include |  CDA dataEnterer CDA dataEnterer | 2005‑09‑07 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 2.16.840.1.113883.10.12.104 | Include |  CDA custodian CDA custodian | 2005‑09‑07 | | 1.3.6.1.4.1.19376.1.3.3.1.4 | Include |  XD-LAB Information Recipient (2017) XD-LAB Information Recipient (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.3.10.2.4 | Include |  XD-LAB LegalAuthenticator (2017) XD-LAB LegalAuthenticator (2017) | 2016‑07‑05 | | 1.3.6.1.4.1.19376.1.8.1.4.3 | Include |  CDA authenticator IHE CDA authenticator IHE | 2016‑07‑09 15:03:59 | | 1.3.6.1.4.1.19376.1.3.3.1.6 | Include |  Ordering Provider (2017) Ordering Provider (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.4.1 | Include |  CDA Participant Specimen Collector CDA Participant Specimen Collector | 2016‑06‑13 14:21:13 | | 1.3.6.1.4.1.19376.1.3.10.2.5 | Include |  CDA Participant Pertinent Insurance Information CDA Participant Pertinent Insurance Information | 2017‑11‑13 16:52:30 | | 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | 2008‑08‑08 | | 2.16.840.1.113883.10.12.113 | Include |  CDA componentOf CDA componentOf | 2005‑09‑07 | | 1.3.6.1.4.1.19376.1.8.1.2.1 | Containment |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 | Containment |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 | Containment |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 | Containment |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.3.10.3.1 | Containment |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.2.5 | Containment |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | 1.3.6.1.4.1.19376.1.8.1.2.6 | Containment |  Procedure Steps Section (2.0) Procedure Steps Section (2.0) | 2014‑05‑13 19:33:12 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.1 (2005‑09‑07) |
|---|
| Example | | example for use case #1 | <ClinicalDocument xsi:schemaLocation="urn:hl7-org:v3 infrastructure/cda/CDA_extended.xsd">
<realmCode code="UV"/> <typeId extension="POCD_HD000040" root="2.16.840.1.113883.1.3"/> <templateId root="1.3.6.1.4.1.19376.1.8.1.1.1"/> <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_1" assigningAuthorityName="IHE PaLM Technical Committee"/> <code code="60568-3" codeSystem="2.16.840.1.113883.6.1" displayName="Pathology Synoptic report"/> <title>Anatomic Pathology Structured Report - Breast Biopsy</title> <effectiveTime value="201001041605-0500"/> <confidentialityCode code="N" displayName="normal" codeSystem="2.16.840.1.113883.5.25"/> <languageCode code="en-US"/> <setId root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008" assigningAuthorityName="IHE PaLM Technical Committee"/> <versionNumber value="1"/> <!-- Patient -->
<recordTarget>
<patientRole>
<id extension="0411886319605719371016" root="1.3.6.1.4.1.19376.1.8.9.2"/> <addr use="HP">
<streetAddressLine>39 East Street</streetAddressLine> <postalCode>69499</postalCode> <city>Appleton</city> <state>WI</state> <country>United States</country> </addr> <telecom nullFlavor="NASK"/> <patient>
<name>
<prefix>Miss</prefix> <given>EVE</given> <family qualifier="BR">ONEWOMAN</family> </name> <administrativeGenderCode code="F" codeSystem="2.16.840.1.113883.5.1"/> <birthTime value="19710921"/> </patient> </patientRole> </recordTarget> <!-- one or more author(s) of the report, with authoring time -->
<author>
<templateId root="1.3.6.1.4.1.19376.1.8.1.4.2"/> <time value="20100104131933-0500"/> <assignedAuthor>
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="801234567897"/> <addr nullFlavor="MSK"/> <telecom value="tel:+33-602030499"/> <assignedPerson>
<name>
<given>Marcel</given> <family>Pathologist</family> <suffix>Ph D</suffix> </name> </assignedPerson> <representedOrganization>
<id root="1.3.6.1.4.1.19376.1.8.9.4" extension="1120456789"/> <name>CANCER INSTITUTE</name> <telecom nullFlavor="MSK"/> <addr nullFlavor="MSK"/> </representedOrganization> </assignedAuthor> </author> <!-- one or more transcriptionists, with transcription time -->
<dataEnterer>
<time value="20100104131720-0500"/> <assignedEntity>
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="A32"/> <addr nullFlavor="MSK"/> <telecom nullFlavor="MSK"/> <assignedPerson>
<name>
<given>Adeline</given> <family>Medsecret</family> </name> </assignedPerson> </assignedEntity> </dataEnterer> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Body) APSR2' (dynamic), not used in use case #1 -->
</informant> <!-- The unique custodian of this document is the sending pathology lab that will administer it (further updates, deprecation) -->
<custodian>
<assignedCustodian>
<representedCustodianOrganization>
<id root="1.3.6.1.4.1.19376.1.8.9.4" extension="1120456789"/> <name>CANCER INSTITUTE</name> <telecom use="PUB" value="0466666666"/> <addr>
<streetAddressLine>38 Cramberry Street</streetAddressLine> <postalCode>69499</postalCode> <city>Appleton</city> <state>WI</state> </addr> </representedCustodianOrganization> </assignedCustodian> </custodian> <!-- One or more additional intended recipients (other than the ordering physician) -->
<informationRecipient>
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.4"/> <intendedRecipient>
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="987"/> <addr>
<streetAddressLine>1600 Clifton Road</streetAddressLine> <city>Atlanta</city> <state>GA</state> <postalCode>30333</postalCode> </addr> <telecom value="tel:404-639-3535"/> <informationRecipient>
<name>
<family>WOULDLIKETOKNOW</family> <given>Thomas</given> </name> </informationRecipient> </intendedRecipient> </informationRecipient> <!-- The unique legal authenticator: The person assuming the final responsibility of the report and signing it -->
<legalAuthenticator>
<time value="20100104152503-0500"/> <signatureCode code="S"/> <assignedEntity>
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="801234567897"/> <assignedPerson>
<name>
<given>Marcel</given> <family>Pathologist</family> </name> </assignedPerson> </assignedEntity> </legalAuthenticator> <!-- Zero or more additional content validator(s): pathologists having validated some part of the report -->
<authenticator>
<templateId root="1.3.6.1.4.1.19376.1.8.1.4.3"/> <time value="20100104142503-0500"/> <signatureCode code="S"/> <assignedEntity>
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="801234567898"/> <addr nullFlavor="MSK"/> <telecom nullFlavor="MSK"/> <assignedPerson>
<name>
<given>Jonas</given> <family>Jones</family> <prefix>MD</prefix> </name> </assignedPerson> </assignedEntity> </authenticator> <!-- The ordering physician -->
<participant typeCode="REF">
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.6"/> <time>
<high value="20091231"/> </time> <associatedEntity classCode="PROV">
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="801234567892"/> <addr nullFlavor="NASK"/> <telecom value="tel:0147150000" use="EC"/> <associatedPerson>
<name>
<prefix>Doctor</prefix> <given>Eva</given> <family>Surgeon</family> <suffix>Ph D</suffix> </name> </associatedPerson> </associatedEntity> </participant> <!-- include template 1.3.6.1.4.1.19376.1.8.1.4.1 'CDA participant specimen collector' (dynamic) 0..* R, not used in use case #1 -->
<!-- include template 1.3.6.1.4.1.19376.1.3.10.2.5 'CDA participant pertinent insurance information' (dynamic) 0..1 R, not used in use case #1 -->
<!-- Identification of the order, extension is Order-ID -->
<inFulfillmentOf>
<order>
<id root="1.3.6.1.4.1.19376.1.8.9.8" extension="12345"/> </order> </inFulfillmentOf> <!-- Documented act(s): The pathology examination procedure, extension is Accession number -->
<documentationOf>
<serviceEvent>
<id root="1.3.6.1.4.1.19376.1.8.9.9" extension="A7102400008"/> <code code="371528001" displayName="Pathology report (record artifact)" codeSystem="2.16.840.1.113883.6.96" codeSystemName="SNOMED CT"/> <lab:statusCode code="completed"/> <effectiveTime>
<!-- Start: Date&time of reception of this order and the attached specimens -->
<low value="200912300922-0500"/> <!-- End -->
<high value="201001041605-0500"/> </effectiveTime> <!-- Performing laboratory -->
<performer typeCode="PRF">
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.7"/> <time>
<high value="201001041605-0500"/> </time> <assignedEntity>
<id root="1.3.6.1.4.1.19376.1.8.9.3" extension="801234567897"/> <representedOrganization>
<id root="1.3.6.1.4.1.19376.1.8.9.4" extension="1120456789"/> <name>CANCER INSTITUTE</name> <telecom nullFlavor="MSK"/> <addr nullFlavor="MSK"/> </representedOrganization> </assignedEntity> </performer> </serviceEvent> </documentationOf> <!-- include template 1.3.6.1.4.1.19376.1.3.1.9999.10.9.16 'RelatedDocument Parent Document' (dynamic) 0..1 R, not used in use case #1 -->
<!-- Patient encounter: The patient stay in the hospital where the surgery was performed -->
<componentOf>
<encompassingEncounter>
<id root="1.3.6.1.4.1.19376.1.8.9.7" extension="234567890"/> <code code="ACUTE" displayName="inpatient acute"/> <effectiveTime>
<high value="2201001040735-0500"/> </effectiveTime> <location typeCode="LOC">
<healthCareFacility classCode="SDLOC">
<id root="1.3.6.1.4.1.19376.1.8.9.4" extension="11223344"/> <serviceProviderOrganization classCode="ORG" determinerCode="INSTANCE">
<name>Surgery theater</name> <asOrganizationPartOf>
<wholeOrganization>
<name>CANCER INSTITUTE</name> </wholeOrganization> </asOrganizationPartOf> </serviceProviderOrganization> </healthCareFacility> </location> </encompassingEncounter> </componentOf> <!-- Structured body -->
<component typeCode="COMP" contextConductionInd="true">
<structuredBody classCode="DOCBODY" moodCode="EVN">
<component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.2.1 'Clinical Information Section' (2014-05-13T14:38:08) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.2.2 'Intraoperative Observation Section' (2014-05-13T19:29:16) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.2.3 'Macroscopic Observation Section' (2014-05-13T11:57:09) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.2.4 'Microscopic Observation Section' (2014-05-13T14:25:17) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.3.10.3.1 'Additionally Specified Observation Section' (2016-11-13T14:28:08) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.2.5 'Diagnostic Conclusion Section' (2014-05-13T19:31:26) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.2.6 'Procedure Steps Section' (2014-05-13T19:33:12) -->
</component> </structuredBody> </component></ClinicalDocument> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | @classCode
|
| cs | 1 … 1 | F | DOCCLIN |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | hl7:templateId
|
| II | 1 … 1 | M | This element is identifying the set of constraints applied to the CDA R2 standard by this IHE specification of a AP report. The following templateId SHALL be present and valued as follows to indicate compliance with the APSR 2.0 content module specification.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.8.1.1.1 |  | hl7:realmCode
|
| CS (extensible) | 1 … 1 | M | This element SHALL be present and is valued from the RealmOfUse [2.16.840.1.113883.1.11.11050] subset, within the VocabularyDomainQualifier value set. In the international context of this profile used as it is without any further extension, the realm code SHALL be <realmCode code="UV"/> (universal). Whenever a national extension has been defined and is used, the realm code SHALL identify this national extension. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | hl7:typeId
|
| II | 1 … 1 | M | This element is a technology-neutral explicit reference to the standard CDA R2. It SHALL be present and valued as follows:
ClinicalDocument/typeId@root = "2.16.840.1.113883.1.3" (which is the OID for HL7 Registered models);
ClinicalDocument.typeId@extension = "POCD_HD000040" (which is the unique identifier for the CDA, Release Two Hierarchical Description).
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @root
|
| uid | 1 … 1 | F | 2.16.840.1.113883.1.3 |  |  | @extension
|
| st | 1 … 1 | F | POCD_HD000040 |  | hl7:id
|
| II | 1 … 1 | M | ClinicalDocument/Id SHALL be present. It represents the unique instance identifier of the clinical document. The combination of the root and extension attributes SHALL provide a globally unique identifier, in accordance with CDA R2, without further constraints.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @root
|
| uid | 1 … 1 | R | Here the OID for PAT exemplary instances, in practice the OID of the LIS |  |  | @extension
|
| st | 1 … 1 | R | Here a hypothetical document ID, most often derived from the accession number
| | | Constraint | A report may have several successive revisions over time, in case corrections or complements are provided by the custodian after the initial release of the report.
The unique id of the current revision of the report is carried by the
id element, and is composed of
- id@root, which SHALL be an OID,
- and optionally id@extension, which can be any string so that the concatenation of the two attributes root and extension provide a globally unique id, which identifies this release of the report.
| | | Example | Report ID from use case #1 <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_1" assigningAuthorityName="IHE PaLM Technical Committee"/> |  | hl7:code
|
| CE (required) | 1 … 1 | M | ClinicalDocument/code SHALL be present. The document type of this content module is always
<code code="60568-3"
codeSystem="2.16.840.1.113883.6.1"
displayName="Pathology Synoptic report"
codeSystemName="LOINC"/>
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @code
|
| CONF | 1 … 1 | F | 60568-3 |  |  | @codeSystem
|
| 1 … 1 | F | 2.16.840.1.113883.6.1 (Logical Observation Identifier Names and Codes) |  |  | @codeSystemName
|
| 1 … 1 | F | LOINC |  |  | @displayName
|
| 1 … 1 | F | Pathology Synoptic report |  | hl7:title
|
| ST | 1 … 1 | M | The APSR <title> SHALL be present. It is the local translation of the code@displayName.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | element content shall be "Anatomic Pathology Structured Report" |
| | | Example | Report title from use case #1 <title>ANATOMIC PATHOLOGY REPORT - BREAST BIOPSY</title> |  | hl7:effectiveTime
|
| TS | 1 … 1 | M | ClinicalDocument/effectiveTime SHALL be present. It contains the creation date & time of the laboratory report as an electronic document. In case this is a new revision replacing a previous version (identified in parentDocument), this is the date & time of the new revision.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | hl7:confidentialityCode
|
| CE (required) | 1 … 1 | M | ClinicalDocument/confidentialityCode SHALL be present in accordance with the HL7 CDA R2 standard.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | The value of @code shall be drawn from value set 2.16.840.1.113883.1.11.16926 HL7 BasicConfidentialityKind (2014‑06‑09) |
|  | hl7:languageCode
|
| CS (required) | 1 … 1 | M | ClinicalDocument/languageCode SHALL be present in accordance with the HL7 CDA R2 standard.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | The value of @code shall be drawn from value set 2.16.840.1.113883.1.11.11526 HumanLanguage (2014‑03‑26) |
|  | hl7:setId
|
| II | 1 … 1 | M | ClinicalDocument/setId SHALL be present to enable further updates of the clinical document. It is an identifier that is common across all revisions of this AP report.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @root
|
| uid | 1 … 1 | R | |  |  | @extension
|
| st | 1 … 1 | R | | | | Constraint | A report may have several successive revisions over time, in case corrections or complements are provided by the custodian after the initial release of the report.
The setId element provides a globally unique identifier that is common across all successive revisions of the report. This identifier is similarly composed of setId@root, which SHALL be an OID, and optionally setId@extension. | | | Example | Report set ID from use case #1 <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008" assigningAuthorityName="IHE PaLM Technical Committee"/> |  | hl7:versionNumber
|
| INT | 0 … 1 | R | ClinicalDocument/versionNumber MAY be present. As requested by the CDA standard, it is an integer value used as versioning.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @value
|
| int | 1 … 1 | R | | | | Constraint | A report may have several successive revisions over time, in case corrections or complements are provided by the custodian after the initial release of the report.
The version number of the current revision of the report is a positive integer (1, 2 …) provided in the versionNumber element. | | | Example | Report version number for use case #1 <versionNumber value="1"/> | | Included | 1 … 1 | M | from 1.3.6.1.4.1.19376.1.3.3.1.1 Human Patient (2017‑06‑07)
The Patient.
The anatomic pathology report is related to ONE SINGLE patient.
-
A patient SHALL be identified with at least one unique patientRole/Id.
-
The patient address and telecom SHALL be provided (or null flavored).
-
The patient identity SHALL provide AT LEASTthe patient full name, sex and date/time of birth.
|  | hl7:recordTarget
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | hl7:patientRole
|
| | 1 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | II | 1 … * | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | AD | 1 … * | | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | TEL | 1 … * | | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | II | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | PN | 1 … * | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  |  | hl7:administrativeGenderCode
|
| CE | 1 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | TS | 1 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | Included | 1 … * | M | from 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11)
The Author Content Module represents an author of the report. This element is repeatable. The sub-element author/time carries the date/time of the authoring action.
At least one ClinicalDocument/author SHALL be present with a time in accordance with the HL7 CDA R2 standard and further constrained by this specification to require the presence of name, addr and telecom. The author/time element carries the date&time the AP report was produced. The AP report can be authored by a software system or by a person or by both.
Source: PaLM Suppl.APSR 2.0‑3: 6.3.6.2
|  | hl7:author
|
| | 1 … * | M | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author |  |  | @typeCode
|
| cs | 1 … 1 | F | AUT | | | Example | author is a person <author typeCode="AUT" contextControlCode="OP">
<time value="201306101654"/> <assignedAuthor classCode="ASSIGNED">
<!-- ... -->
</assignedAuthor></author> |  |  | hl7:templateId
|
| | 1 … 1 | M | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.8.1.4.2 |  |  | hl7:time
|
| TS | 1 … 1 | M | The authoring time is the date & time that this author contributed to the document. It SHALL be provided. | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author |  |  | hl7:assignedAuthor
|
| | 0 … 1 | C | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | cs | 0 … 1 | F | ASSIGNED | | II | 1 … * | M | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | AD | 1 … * | M | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | TEL | 1 … * | M | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | Choice | 1 … 1 | | The author is either an assigned person or an authoring device.
Elements to choose from:- hl7:assignedPerson
- hl7:assignedAuthoringDevice
| | | 0 … 1 | C | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | Included | 0 … * | | from 2.16.840.1.113883.10.12.152 CDA Person (2005‑09‑07)
The name(s) SHALL be given. | | | 0 … 1 | F | PSN | | | 0 … 1 | F | INSTANCE | | PN | 0 … * | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author |  |  |  |  | hl7:assignedAuthoringDevice
|
| | 0 … 1 | C | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | cs | 0 … 1 | F | DEV | | cs | 0 … 1 | F | INSTANCE |  |  |  |  |  | hl7:manufacturerModelName
|
| SC | 0 … 1 | R | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | SC | 0 … 1 | R | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | CE | 0 … 1 | R | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author |  |  |  | hl7:representedOrganization
|
| | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | | Example | represented organization for author as a device <representedOrganization classCode="ORG" determinerCode="INSTANCE">
<name>
<!-- ... -->
</name></representedOrganization> | | Included | 0 … 1 | | from 2.16.840.1.113883.10.12.151 CDA Organization (2005‑09‑07)
The identifier(s) SHOULD, the name SHALL, the telecom(s) and the address(es) MAY be given. | | | 0 … 1 | F | ORG | | | 0 … 1 | F | INSTANCE | | II | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | ON | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | TEL | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | AD | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author |  |  |  |  | hl7:standardIndustryClassCode
|
| CE | 0 … 1 | | SHALL be chosen from domain OrganizationIndustryClass | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | | 0 … 1 | F | PART | | II | 0 … * | R | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | CE | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | CONF | 0 … 1 | F | 2.16.840.1.113883.5.111 (Role Code) | | CS | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | | CONF | | The value of @code shall be drawn from value set 2.16.840.1.113883.1.11.15999 RoleStatus (DYNAMIC) |
| | IVL_TS | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | | 0 … 1 | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | | 0 … 1 | F | ORG | | | 0 … 1 | F | INSTANCE | | II | 0 … * | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | ON | 0 … * | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | TEL | 0 … * | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | AD | 0 … * | | | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author |  |  |  |  |  |  | hl7:standardIndustryClassCode
|
| CE | 0 … 1 | | SHALL be chosen from domain OrganizationIndustryClass | PaLM Suppl.APSR 2.0‑3:6.3.6.2 Author | | Included | 0 … * | R | from 2.16.840.1.113883.10.12.103 CDA dataEnterer (2005‑09‑07)
Transcriptionist
|  | hl7:dataEnterer
|
| | 0 … * | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| | 0 … 1 | F | ENT |  |  | @contextControlCode
|
| | 0 … 1 | F | OP |  |  | hl7:time
|
| TS | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | hl7:assignedEntity
|
| | 1 … 1 | | Contains 2.16.840.1.113883.10.12.153 CDA AssignedEntity (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | hl7:informant
|
| | 0 … * | R | Zero or more informant MAY be mentioned in the header of the report. An informant is either an assignedEntity (a professional participating to the healthcare process, and who was assigned a defined role in that process) or a relatedEntity (a person who knows the patient and has provided relevant information concerning the patient). Hence the condition is either assignedEntity is present or relatedEntity is present. These two elements are defined in the content module “Informant”.
Source: PaLM Suppl. APSR 2.0-3: 6.3.6.5
Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| | | Included | 1 … 1 | M | from 2.16.840.1.113883.10.12.104 CDA custodian (2005‑09‑07)
ClinicalDocument/custodian SHALL be present with an id in accordance with the HL7 CDA R2 standard and further constrained by this specification to require the presence of name, addr and telecom. It represents the organization that is in charge of maintaining the AP report.
|  | hl7:custodian
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| | 0 … 1 | F | CST |  |  | hl7:assignedCustodian
|
| | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | F | ASSIGNED |  |  |  | hl7:representedCustodianOrganization
|
| | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | F | ORG | | | 0 … 1 | F | INSTANCE | | II | 1 … * | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | ON | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | TEL | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | AD | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | Included | 0 … * | R | from 1.3.6.1.4.1.19376.1.3.3.1.4 XD-LAB Information Recipient (2008‑08‑08)
The Intended Recipient Content Module represents a healthcare provider,
other than the ordering physician, expecting to receive a copy of the report. This repeatable element informationRecipient of the CDA header is used to list the intended recipients who were known at the time the report was created and issued.
Source: PaLM TF-3: 6.3.2.14
|  | hl7:informationRecipient
|
| | 0 … * | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | hl7:templateId
|
| II | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.3.1.4 |  |  | hl7:intendedRecipient
|
| | 1 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | II | 0 … * | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | AD | 1 … * | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | TEL | 1 … * | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | hl7:informationRecipient
|
| | 0 … 1 | | Contains 1.3.6.1.4.1.19376.1.3.10.9.18 PlayingEntity or person with Name (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | hl7:receivedOrganization
|
| | 0 … 1 | | Contains 1.3.6.1.4.1.19376.1.3.10.9.13 Organization with Name, Addr, Telecom (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | Included | 1 … 1 | M | from 1.3.6.1.4.1.19376.1.3.10.2.4 XD-LAB LegalAuthenticator (2016‑07‑05)
The Legal authenticator Content Module describes a pathologist having verified the content of the report, using the element legalAuthenticator.
The report SHALL have one legal Authenticator
Source: PaLM TF-3: 6.3.2.15
|  | hl7:legalAuthenticator
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | hl7:time
|
| TS | 1 … 1 | R | The sub-element time carries the date&time this legal authentication took place. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | hl7:signatureCode
|
| CS | 1 … 1 | R | The sub-element signatureCode carries the “signed” (S) status | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | CONF | 0 … 1 | F | S |  |  | hl7:assignedEntity
|
| | 1 … 1 | R | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | AD | 1 … * | R | Constrained by this specification to require the presence of name, addr and telecom. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | TEL | 1 … * | R | Constrained by this specification to require the presence of name, addr and telecom. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom.
Contains 1.3.6.1.4.1.19376.1.3.10.9.18 PlayingEntity or person with Name (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | hl7:representedOrganization
|
| | 0 … 1 | | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom.
Contains 1.3.6.1.4.1.19376.1.3.10.9.13 Organization with Name, Addr, Telecom (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | Included | 0 … * | R | from 1.3.6.1.4.1.19376.1.8.1.4.3 CDA authenticator IHE (2016‑07‑09 15:03:59)
The Content validator Content Module describes a pathologist having verified the content of the report, using the element authenticator. This element authenticator is used when the pathologist having verified the content of the report is distinct from the pathologist assuming the legal responsibility for this report, described in the legalAuthenticator element.
The report MAY have zero or more Content Validators.
Source: PaLM Suppl. APSR 2.0-3: 6.3.6.3
|  | hl7:authenticator
|
| | 0 … * | R | | PaLM Suppl. APSR 2.0‑3: 6.3.6.3 Content validator |  |  | @typeCode
|
| cs | 1 … 1 | F | AUTHEN |  |  | hl7:templateId
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.6.3 Content validator | | uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.8.1.4.3 |  |  | hl7:time
|
| TS | 1 … 1 | R | Time of validation | PaLM Suppl. APSR 2.0‑3: 6.3.6.3 Content validator |  |  | hl7:assignedEntity
|
| | 0 … 1 | C | AssignedPerson SHALL be given with name, representedOrganization MAY be given.
Contains 2.16.840.1.113883.10.12.153 CDA AssignedEntity (2005‑09‑07) | PaLM Suppl. APSR 2.0‑3: 6.3.6.3 Content validator | | Included | 1 … 1 | R | from 1.3.6.1.4.1.19376.1.3.3.1.6 Ordering Provider (2008‑08‑08)
The Ordering Provider Content Module represents the physician who has submitted the specimen examination order to the anatomic pathology laboratory. As specified in PaLM TF-3, this physician is represented in the CDA header as a participant element with the typeCode attribute valued “REF”. The sub-element participant/time carries the date/time of issuance of the
order.
Source: PaLM TF-3: 6.3.2.17
|  | hl7:participant
|
| | 1 … 1 | R | Referral Ordering Physician | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| cs | 1 … 1 | F | REF |  |  | hl7:templateId
|
| II | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.3.1.6 |  |  | hl7:time
|
| IVL_TS | 1 … 1 | R | This element represents the date and time the order was placed. Time MAY be present. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | hl7:associatedEntity
|
| | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | AD | 1 … * | R | The address of this person (referral ordering physician) SHALL be present. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | TEL.AT | 1 … * | R | The telecom of this person (referral ordering physician) SHALL be present. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | Schematron assert | role |  error error | | | | test | not(hl7:assignedPerson) or hl7:assignedPerson/hl7:name | | | | Message | The <name> sub-element SHALL be present when <assignedPerson> present. | | | | 0 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | Included | 0 … * | R | from 1.3.6.1.4.1.19376.1.8.1.4.1 CDA Participant Specimen Collector (2016‑06‑13 14:21:13)
The Specimen Collector Content Module is only used in case a specimen
provided as input to the AP act documented in this report, was collected by a different party than the ordering physician. In that case, this specimen collector is represented in the CDA header as a participant element with the typeCode attribute valued “DIST” and the sub-element participant/time carries the time period of the specimen collection.
Source: PAT TF-3: 6.2.6.1
|  | hl7:participant
|
| | 0 … * | R | The Specimen Collector Content Module is only used in case a specimen provided as input to the AP act documented in this report, was collected by a different party than the ordering physician. In that case, this specimen collector is represented in the CDA header as a participant element with the typeCode attribute valued “DIST” and the sub-element participant/time carries the time period of the specimen collection.
| PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector |  |  | @typeCode
|
| cs | 1 … 1 | F | DIST |  |  | hl7:templateID
|
| II | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.8.1.4.1 |  |  | hl7:time
|
| IVL_TS | 1 … 1 | M | Specimen collection time
The specimen collection time is an interval, which may be reduced to a point in time (see usage of data type IVL_TS).
| PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector |  |  | hl7:associatedEntity
|
| | 0 … 1 | R | At least one of the two elements
associatedPerson
and
scopingOrganization
must be present. Both may be present.
| PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | cs | 1 … 1 | F | PROV | | II | 1 … * | R | | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | AD | 1 … * | R | | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | TEL.AT | 1 … * | R | | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | | 0 … * | C | Person who collected the specimen.
Only full name(s) SHALL be given.
Contains 2.16.840.1.113883.10.12.152 CDA Person (2005‑09‑07) | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | | 0 … 1 | C | Organization taking care for specimen collection.
Identifiers, name, telecom, and address SHOULD be given.
Contains 2.16.840.1.113883.10.12.151 CDA Organization (2005‑09‑07) | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector | | Included | 0 … 1 | R | from 1.3.6.1.4.1.19376.1.3.10.2.5 CDA Participant Pertinent Insurance Information (2017‑11‑13 16:52:30)
Key insurance information of the patient |  | hl7:participant
|
| | 0 … 1 | R | Insurance information of the patient in context for reporting to cancer registries (in Germany) | PaLM Suppl. 2.0‑3:6.3.6.13 |  |  | @typeCode
|
| cs | 1 … 1 | F | HLD |  |  | hl7:templateId
|
| II | 1 … * | M | | PaLM Suppl. 2.0‑3:6.3.6.13 | | uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.10.2.5 |  |  | hl7:time
|
| IVL_TS | 0 … 1 | R | time shows the end of the current insurance period, e.g. end of the quartal | PaLM Suppl. 2.0‑3:6.3.6.13 | | | Example | <time>
<high value="20171231"/></time> |  |  | hl7:associatedEntity
|
| | 1 … 1 | M | Data of the insured person | PaLM Suppl. 2.0‑3:6.3.6.13 | | cs | 1 … 1 | F | POLHOLD | | II | 0 … * | R | ID(s) of the insured person (patient), e.g. ID of the insured, ID of the health card, etc. | PaLM Suppl. 2.0‑3:6.3.6.13 | | CE | 0 … 1 | R | status of the insured person | PaLM Suppl. 2.0‑3:6.3.6.13 | | CONF | 0 … 1 | F | 2.16.840.1.113883.5.111 (Role Code) | | | Example | <code code="SELF" codeSystem="2.16.840.1.113883.5.111" displayName="self">
<translation code="1" codeSystem="2.16.840.1.113883.3.7.1.1" displayName="member"/></code> | | CV | 0 … 1 | R | further codes of the status of the insured person,
e.g. for Germany from value set S_KBV_VERSICHERTENSTATUS
1.2.276.0.76.11.162 | PaLM Suppl. 2.0‑3:6.3.6.13 | | AD | 0 … 1 | R | | PaLM Suppl. 2.0‑3:6.3.6.13 | | TEL | 0 … * | R | | PaLM Suppl. 2.0‑3:6.3.6.13 | | | 0 … 1 | R | Data of the insured person, which may be different from the patient.
Contains 2.16.840.1.113883.10.12.152 CDA Person (2005‑09‑07) | PaLM Suppl. 2.0‑3:6.3.6.13 | | | 1 … 1 | R | Data of the Insurance company, including the ID, the address data and the name.
In Germany the ID is formed by the ID itself (@extension = so-called IKNR) and the OID for IKNR (@root="1.2.276.0.76.4.5") in Germany.
Contains 2.16.840.1.113883.10.12.151 CDA Organization (2005‑09‑07) | PaLM Suppl. 2.0‑3:6.3.6.13 |  | hl7:inFulfillmentOf
|
| | 0 … 1 | R | The inFulfillmentOf/order element MAY be present. It represents the Placer Order or the Placer Group that was fulfilled, the Id of which is carried by inFulfillmentOf/order/id.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| cs | 1 … 1 | F | FLFS |  |  | hl7:order
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | cs | 1 … 1 | F | ACT | | cs | 1 … 1 | F | RQO | | II | 1 … * | R | Placer order group ID or Placer order ID
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | hl7:documentationOf
|
| | 1 … 1 | M | Documented act. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| cs | 1 … 1 | F | DOC |  |  | hl7:serviceEvent
|
| | 1 … 1 | M | The AP report is documenting a service (documentationOf/serviceEvent) performed by a surgical pathology laboratory. The Laboratory Performer Content Module represents this laboratory, and is fully described in the sub-element
documentationOf/serviceEvent/performer.
In case more than one laboratory contributed to a service, only the primary laboratory is in the CDA header, attached to the serviceEvent element, and the other (secondary) laboratories are described only in the sections of the report that they contributed to, in the body of the report.
Source: PaLM TF3: 6.3.2.19
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | cs | 1 … 1 | R | | | cs | 1 … 1 | R | | | II | 1 … * | M | ID of the Act, i.e. Accession number | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | CE (extensible) | 0 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | @code shall be "PATREPE" | | @codeSystem shall be "2.16.840.1.113883.5.4" | | @codeSystemName shall be "HL7 ActCode" | | @displayName shall be "pathology report entry task" | | or | | @code shall be "371528001" | | @codeSystem shall be "2.16.840.1.113883.6.96" | | @codeSystemName shall be "SCT" | | @displayName shall be "Pathology report (record artifact)" |
| | IVL_TS | 0 … 1 | R | Use of sub element documentationOf/serviceEvent/effectiveTime to document the time boundaries of events in the document is appropriate.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | CS (required) | 0 … 1 | R |
The statusCode below documentationOf/serviceEvent is an extension to the CDA R2 standard, added by PaLM TF-3 to distinguish a preliminary report (statusCode@code="active") from a final report (statusCode@code="completed").
The statusCode sub element is further described in section A.3 of PaLM TF-3. This sub-element is required. When it is not there, the documented Act is assumed to be completed and the report is assumed to be a final report.
This extension to the standard is protected by a dedicated namespace associated in the ClinicalDocument element to the prefix lab:
<Clinical Document xmlns:lab="urn:oid:1.3.6.1.4.1.19376.1.3.2" … >
Source: PaLM TF3: 6.3.2.19
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | Constraint | The statusCode@code="completed" is only applicable, if ALL content modules of the Clinical Document have the statusCode@code="completed", too, or have the statusCode@code="aborted".
| | | CONF | | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.4 ActStatusActiveCompleted (2008‑08‑08) |
| | | Schematron assert | role |  error error | | | | test | hl7:serviceEvent/lab:statusCode[@code='completed'] | | | | Message | Status code of all Content Modules SHALL be "complete" or "aborted" | | | | 0 … * | R | Contains 1.3.6.1.4.1.19376.1.3.3.1.7 Laboratory Performer (2008‑08‑08) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.7']] |
| |  | hl7:relatedDocument
|
| | 0 … 1 | R | ClinicalDocument/relatedDocument/parentDocument
This element SHALL be present in case of an update replacement of a previous report. In this case relatedDocument@typeCode attribute SHALL be valued "RPLC", the new report replacing the parent one.
Note 1: A non-final AP structured report published in an XDS infrastructure will likely be replaced afterwards by the final report. When this event occurs, the Content Creator Actor SHALL apply the following rules:
- ClinicalDocument/setId SHALL have the same value in the new report as in the replaced report.
- ClinicalDocument/versionNumber SHALL be incremented in the replacing report (i.e., the final one).
- ClinicalDocument/relatedDocument@typeCode attribute SHALL be valued ”RPLC”
- ClinicalDocument/relatedDocument/parentDocument/id in the new report SHALL be equal to ClinicalDocument/ id of the replaced document.
The Document Source Actor SHALL apply the following rules on XDSDocumentEntry metadata:
- The final report SHALL be associated with the previously published one, using RPLC relationship and the previous report SHALL be “Deprecated” as described in ITI TF-2:4.1.6.1.
Note 2: A non-final report can also be replaced by a more recent, albeit still non-final report. The rules above also apply in this case.
Note 3: A final report can also be replaced by a corrective final report. The rules above also apply in this case. | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| cs | 1 … 1 | F | RPLC | | | relatedDocument@typeCode attribute SHALL be valued "RPLC"
|  |  | hl7:parentDocument
|
| ANY | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | II | 1 … 1 | M | SHALL be equal to ClinicalDocument/ id of the replaced document.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | II | 1 … 1 | M | SHALL have the same value in the new report as in the replaced report.
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | INT | 0 … 1 | R | SHALL have the same value as in the replaced report (when provided there).
| PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | Included | 0 … 1 | R | from 2.16.840.1.113883.10.12.113 CDA componentOf (2005‑09‑07)
The ClinicalDocument/componentOf/encompassingEncounter element MAY be present.
It describes the encounter during which the reported AP observations were ordered. When present the encounter SHALL:
- be identified with an id element: encompassingEncounter/id
- The encounter SHALL have an effective time that represents the time interval (possibly still running, e.g., an inpatient current stay) of the encounter or a point in time at which the encounter took place (e.g., an outpatient consultation): encompassingEncounter/ effectiveTime
The encounter MAY provide any number of encounter participants (encompassingEncounter/encounterParticipant/assignedEntity). When present, encounter participants SHALL be in accordance with the HL7 CDA R2 standard with a time and further constrained by this specification to require the presence of name, addr and telecom. Additionally, the encounter participant SHALL have a typeCode with one the values selected from the x_EncounterParticipant domain: The encounter MAY precise the patient location during this encounter. This is the healthcare facility in which the patient was located when the reported AP observations were ordered: encompassingEncounter/location/healthCareFacility. This healthcare facility can be represented as a physical place (e.g., room, floor, building, office) or as an organization (e.g., service, department, team) or both: healthCareFacility/location, healthCareFacility/serviceProviderOrganization. |  | hl7:componentOf
|
| | 0 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| | 0 … 1 | F | COMP |  |  | hl7:encompassingEncounter
|
| | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | F | ENC | | | 0 … 1 | F | EVN | | II | 0 … * | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | CE | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | The value of @code shall be drawn from value set 2.16.840.1.113883.1.11.13955 ActEncounterCode (DYNAMIC) |
| | IVL_TS | 1 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | hl7:dischargeDispositionCode
|
| CE | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | shall be drawn from concept domain "EncounterDischargeDisposition" |
| | | 0 … 1 | | Contains 2.16.840.1.113883.10.12.153 CDA AssignedEntity (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | F | RESP | | | 0 … * | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | cs | 1 … 1 | R | | | | CONF | | The value of @typeCode shall be drawn from value set 2.16.840.1.113883.1.11.19600 x_EncounterParticipant (DYNAMIC) |
| | IVL_TS | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 1 … 1 | | Contains 2.16.840.1.113883.10.12.153 CDA AssignedEntity (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | F | LOC | | | 1 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | 0 … 1 | F | SDLOC | | II | 0 … * | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | CE | 0 … 1 | | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | | CONF | | The value of @code shall be drawn from value set 2.16.840.1.113883.1.11.17660 ServiceDeliveryLocationRoleType (DYNAMIC) |
| | | 0 … 1 | | Contains 2.16.840.1.113883.10.12.317 CDA Place (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  |  |  | hl7:serviceProviderOrganization
|
| | 0 … 1 | | Contains 2.16.840.1.113883.10.12.151 CDA Organization (DYNAMIC) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  | hl7:component
|
| | 1 … 1 | R | Body of the CDA Clinical document | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 1 … 1 | F | true |  |  | hl7:structuredBody
|
| | 1 … 1 | R | | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module | | cs | 1 … 1 | F | DOCBODY | | cs | 1 … 1 | F | EVN | | | 0 … 1 | R | The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, clinical laboratory data, specimen(s) description, collection procedure, reason for anatomic pathology procedure.
Contains 1.3.6.1.4.1.19376.1.8.1.2.1 Clinical Information Section (2014‑05‑13 14:38:08) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.2.1']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true | | | 0 … 1 | R |
Contains 1.3.6.1.4.1.19376.1.8.1.2.2 Intraoperative Observation Section (2014‑05‑13 19:29:16) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.2.2']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true | | | 0 … 1 | R | The Macroscopic Observation section contains the description of the specimen received or obtained by the laboratory (specimen type and state), the gross observation, links to gross images, if taken, processing information and tissue disposition (representative sampling and tissue submitted for additional studies or sent to biorepository.
Contains 1.3.6.1.4.1.19376.1.8.1.2.3 Macroscopic Observation Section (2014‑05‑13 11:57:09) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.2.3']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true | | | 0 … 1 | R |
The Microscopic Observation section contains optionally the histopathologic findings of the case and many laboratories use this section to record the results of histochemical and immunohistochemical stains.
Contains 1.3.6.1.4.1.19376.1.8.1.2.4 Microscopic Observation Section (2014‑05‑13 14:25:17) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.2.4']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true | | | 0 … 1 | R | The Additional Specific Observation section includes additional pathologic finding(s) and the results of ancillary study(ies) with non-morphological methods (e.g. flow cytometry, cytogenetics, molecular pathology, etc.) and may include diagrams and still images or virtual slides, if taken.
Contains 1.3.6.1.4.1.19376.1.3.10.3.1 Additional Specified Observation Section (2016‑11‑13 14:28:08) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.10.3.1']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true | | | 1 … 1 | M |
The Diagnostic Conclusion section contains diagnoses on all specimens that are delivered to the pathology department from one operation or patient visit to a single clinician on a particular day. The diagnoses for each specimen or group of specimens are reported separately. This section includes additional pathologic finding(s) and the results of ancillary study(ies) and may include diagrams and still images or virtual slides, if taken. In case of cancer, this section includes the cancer checklist.
Contains 1.3.6.1.4.1.19376.1.8.1.2.5 Diagnostic Conclusion Section (2014‑05‑13 19:31:26) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.2.5']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true | | | 0 … 1 | R |
The Procedure steps section contains the description of tissue dissection: representative specimens and derived specimens dissected for other ancillary procedures (flow cytometry, cytogenetics, molecular studies, electron microscopy, etc.) or biorepository.
Contains 1.3.6.1.4.1.19376.1.8.1.2.6 Procedure Steps Section (2014‑05‑13 19:33:12) | PaLM Suppl. APSR 2.0‑3: 6.3.1.1 APSR clinical document content module |  |  |  | where [@typeCode='COMP'] [hl7:section [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.2.6']]] |
| | | cs | 1 … 1 | F | COMP | | bl | 1 … 1 | F | true |
|
6.3.1.2.1 General constraints that apply to APSR
- When a section has a text element and one or more entry element, the content coded for machine-processing in the entries SHALL be completely transcribed into human-readable content in the text element.
- Conversely the text element MAY contain pieces of information, which are not available in machine-readable format in any entry element of the section.
- Information that is sent SHALL clearly identify distinctions between:
None
It is known with complete confidence that there are none. This indicates that the sender knows that there is no relevant information of this kind that can be sent.
None Known
None known at this time, but it is not known with complete confidence that none exist.
Asked but unknown
The information was requested but could not be obtained. Used mainly in the context where an observation was made but the result could not be determined.
Unknown
The information is not known, or is otherwise unavailable.
Other, not specified
The actual value does not belong to the assigned value set and is not reported at all by the author.
Other, specify
The actual value does not belong to the assigned value set and the author of the report provides this foreign value anyway.
Not applicable
No proper value is applicable in this context.
Sections that are required to be present but have no information should use one of the above phrases where appropriate in the text element.
Structural elements that are required but have no information shall provide a “nullFlavor” attribute coding the reason why the information is missing.
| Situation |
nullFlavor |
HL7 Definition
|
| Asked but unknown |
ASKU
|
Information was sought but not found
|
| Unknown
|
UNK |
A proper value is applicable, but not known
|
| Other, not specified
|
OTH |
The actual value is not an element in the value domain of a variable. (e.g., concept not provided by required code system).
|
| Not applicable
|
NA |
No proper value is applicable in this context
|
| Temporarily not available
|
NAV |
Information is not available at this time but it is expected that it will be available later.
|
The two situations “None” and “None known” represent effective values, which are part of the related value sets.
The situation “Other, specify” can be handled in two ways in a coded data element:
- Leaving empty the code attribute and providing the non-coded answer in the originalText attribute.
- Providing a value coded from a different coding scheme, when the coding strength of the element is “CWE” (coded with extensions). The attributes code, displayName, codeSystem and codeSystemName then describe the foreign code.
For ancillary techniques, the situation “ not performed” or “none performed” is represented by nullFlavor = NAV.
6.3.1.2.2 Common structure for CDA APSR
Figure 6.3.1.2.2-1 summarizes the common structure of the first six CDA APSR Section Content Modules specified here. Regarding the machine-readable part, the figure highlights the organization of entries within a section and of observations within an entry. Specific details such as the structure of sub-sections are not shown on this global picture.
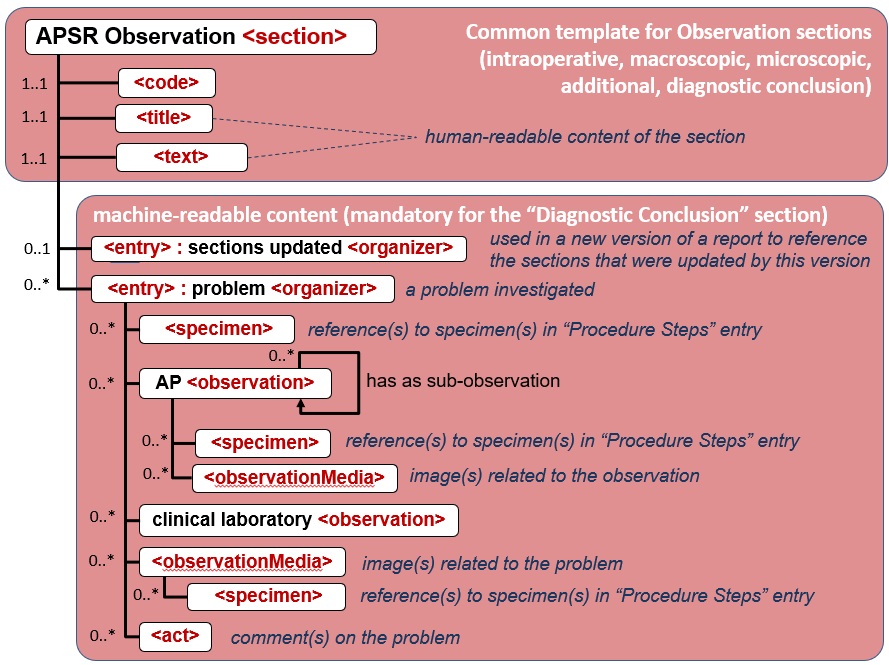
Figure 6.3.1.2.2-1 CDA APSR: common structure of machine-readable content for CDA APSR Section Content Modules except Procedure Step Content Module
Note 1: In order to facilitate a further de-identification process of CDA AP reports for some secondary use (biosurveillance, epidemiology…) the producer of an APSR SHOULD avoid populating any patient identification data (name, sex, birthdate, address …) into the body of the report (neither <entry> elements nor <text> elements). The appropriate location for patient identification data is the CDA header exclusively.
Note 2: The AP sections are those shown on figure 10.4.1-1 of Volume 1.
Note 3: Possible sub sections are shown on figure 10.4.1-1 of Volume 1.
Figure 6.3.1.2.2-2 shows the common structure of the Procedure Step Content Module specified here, too.
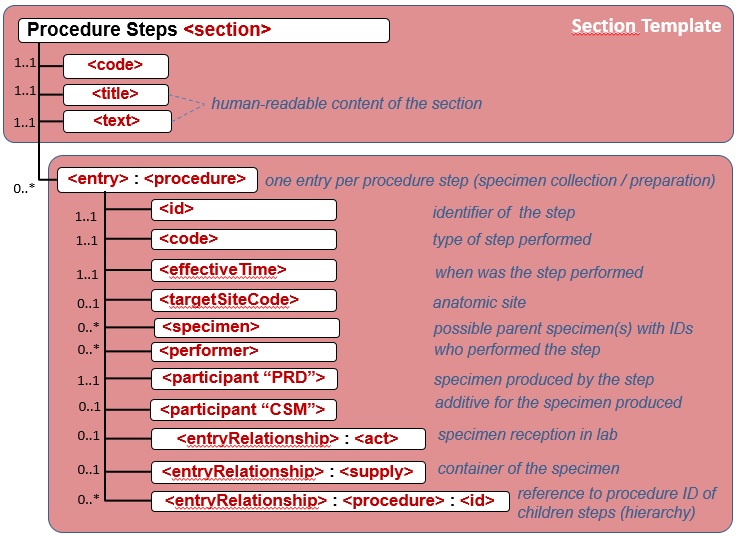
Figure 6.3.1.2.2-2 CDA APSR: common structure of machine-readable content for Procedure Step Content Module
6.3.2 Place Holder
Intentionally left blank
6.3.3 Place Holder
Intentionally left blank
6.3.4 CDA R2 <section> Content Modules
6.3.4.1 Clinical Information <section> - 1.3.6.1.4.1.19376.1.8.1.2.1
6.3.4.1.1 Definition and Purpose
The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, reason for anatomic pathology procedure, clinical laboratory data, specimen collection procedure including target site, performer, specimen type, specimen(s) clinical description, and tumor site in case of a cancer. The only entry for this section is the Problem Organizer Entry module.
6.3.4.1.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.8.1.2.1 | Effective Date | valid from 2014‑05‑13 14:38:08 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ClinicalInformationSection | Display Name | Clinical Information Section |
|---|
| Description | The Clinical Information section contains the information provided by the ordering physician: Clinical history, preoperative diagnosis, postoperative diagnosis, reason for anatomic pathology procedure, clinical laboratory data, information about specimens collected in this case (including target site, performer, specimen type, specimen(s) clinical description, and tumor site in case of a cancer). |
|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.8.1.2.1 |
|---|
| Label | PaLM Suppl. 2.0‑3: 6.3.4.1 Clinical information section
|
|---|
| Classification | CDA Section Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 1 template, Uses 5 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.1.1 | Containment |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.3.6 | Containment |  Problem Organizer (2.0) Problem Organizer (2.0) | 2015‑08‑13 10:24:55 | | 1.3.6.1.4.1.19376.1.8.1.4.2 | Include |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.5.3.1.3.1 | Containment |  IHE Reason for Referral Section (2014) IHE Reason for Referral Section (2014) | 2016‑09‑26 07:58:18 | | 1.3.6.1.4.1.19376.1.5.3.1.3.4 | Containment |  IHE History of Present Illness Section (2014) IHE History of Present Illness Section (2014) | 2013‑01‑31 | | 1.3.6.1.4.1.19376.1.5.3.1.3.6 | Containment |  IHE Active Problems Section (2014) IHE Active Problems Section (2014) | 2015‑10‑05 16:02:07 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.201 (2005‑09‑07) |
|---|
| Example | | generated example | <section classCode="DOCSECT" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.8.1.2.1"/> <code code="22636-5" codeSystem="2.16.840.1.113883.6.1" displayName="Pathology report relevant history"/> <title>CLINICAL INFORMATION SECTION</title> <text/> <languageCode/> <entry xsi:type="ANY" typeCode="COMP" contextConductionInd="true">
<!-- template 1.3.6.1.4.1.19376.1.8.1.3.6 'Problem Organizer' (2015-08-13T10:24:55) -->
</entry> <!-- include template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) 0..* C -->
<component>
<!-- template 1.3.6.1.4.1.19376.1.5.3.1.3.1 'IHE Reason for Referral Section' (2016-09-26T07:58:18) -->
</component> <component>
<!-- template 1.3.6.1.4.1.19376.1.5.3.1.3.4 'History of Present Illness Section' (2013-01-31T00:00:00)
-->
</component> <component>
<!-- template 1.3.6.1.4.1.19376.1.5.3.1.3.6 'IHE Active Problems Section' (2015-10-05T16:02:07) -->
</component></section> |
|
|
6.3.4.2 Intraoperative Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.2
6.3.4.2.1 Definition and Purpose
The Intraoperative Observation section contains an intraoperative diagnosis for each specimen examined, the specimen identification and description, intraoperative observation procedure description (frozen section, gross examination, intraoperative cytology) and derived specimen dissected for other ancillary procedures (flow cytometry, cytogenetics, molecular studies, and electron microscopy). The only entry for this section is the Problem Organizer Entry module.
6.3.4.2.2 Specification and Example
6.3.4.3 Macroscopic Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.3
6.3.4.3.1 Definition and Purpose
The Macroscopic Observation section contains the description of the specimen(s) received or obtained by the laboratory (specimen type and state), the gross observation, links to gross images, if taken, processing information and tissue disposition (representative sampling and tissue submitted for additional studies or sent to biorepository. The only entry for this section is the Problem Organizer Entry module.
6.3.4.3.2 Specification and Example
6.3.4.4 Microscopic Observation <section> - 1.3.6.1.4.1.19376.1.8.1.2.4
6.3.4.4.1 Definition and Purpose
The Microscopic Observation section contains optionally the histopathologic findings of the case and many laboratories use this section to record the results of histochemical and immunohistochemical stains.The only entry for this section is the Problem Organizer Entry module.
6.3.4.4.2 Specification and Example
6.3.4.5 Additional Specified Observation <section> - 1.3.6.1.4.1.19376.1.3.10.3.1
6.3.4.5.1 Definition and Purpose
The Additional Specified Observation section includes additional pathologic finding(s) and the results of ancillary studies with non-morphological methods (e.g. flow cytometry, cytogenetics, molecular pathology, etc.) and may include diagrams and still images or virtual slides, if taken. Further CDA content section modules as subsections are allowed. The only entry for this section is the Problem Organizer Entry module.
6.3.4.5.2 Specification and Example
6.3.4.6 Diagnostic Conclusion <section> - 1.3.6.1.4.1.19376.1.8.1.2.5
6.3.4.6.1 Definition and Purpose
The Diagnostic Conclusion section contains diagnoses on all specimens that are delivered to the pathology department from one operation or patient visit to a single clinician on a particular day. The diagnoses for each specimen sample or group of specimen samples are reported separately. This section may include diagrams and still images or virtual slides, if taken. In case of cancer, this section may also include a cancer checklist. The only mandatory entry for this section is the Problem Organizer Entry module, additionally the Update Organizer Entry module MAY be used.
6.3.4.6.2 Specification and Example
6.3.4.7 Procedure steps <section> - 1.3.6.1.4.1.19376.1.8.1.2.6
6.3.4.7.1 Definition and Purpose
The Procedure steps section contains the description of tissue sampling and dissection: representative specimens and derived specimens dissected for other ancillary procedures (flow cytometry, cytogenetics, molecular studies, electron microscopy, etc.) or biorepository. The only entry for this section is the Specimen Procedure Steps Entry module.
6.3.4.7.2 Specification and Example
6.3.5 CDA R2 <entry> Content Modules
6.3.5.1 Common Specification for all APSR Entry Content Modules
The unique <entry> Content Module available in all the sections except Procedure Steps <section> of the APSR template is the Problem Organizer <entry>.
The unique <entry> Content Module available in Procedure Steps <section> of the APSR template is Specimen Procedure Step <entry>.
Each <entry> Content Module carries machine-readable data related to one Problem on one specimen or on a group of specimens observed for this section.
Each <entry> Content Module is repeatable below its section, so as to support the use cases where a section reports on more than one problem on one specimen or one group of specimens.
6.3.5.2 Problem Organizer <entry> – 1.3.6.1.4.1.19376.1.8.1.3.6
6.3.5.2.1 Definition and purpose
This Content Module is usable as only <entry> within any Section module except Procedure steps <section>.
The problem organizer groups the battery of observations together with embedded images performed to investigate on a single problem identified on a specimen or on a group of specimens. If a Problem cannot be addressed specifically the Problem organizer allows also the grouping around one specimen. The problem type is coded by using a component with a generic CDA observation, not to confuse with the series of AP and other observations organized by this Organizer content module.
6.3.5.2.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.8.1.3.6 | Effective Date | valid from 2015‑08‑13 10:24:55 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ProblemOrganizer | Display Name | Problem Organizer |
|---|
| Description | Template CDA Organizer (prototype, directly derived from POCD_RM000040 MIF)
The problem organizer groups the battery of observations performed to investigate on a single problem identified on a specimen or group of specimens.
|
|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.8.1.3.6 |
|---|
| Label | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer
|
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 7 templates, Uses 11 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.3.1 | Containment |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | 1.3.6.1.4.1.19376.1.8.1.2.1 | Containment |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 | Containment |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 | Containment |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 | Containment |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.8.1.2.5 | Containment |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 1.3.6.1.4.1.19376.1.8.1.4.10 | Containment |  Embedded Image IHE Embedded Image IHE | 2016‑06‑22 16:08:01 | | 2.16.840.1.113883.10.12.307 | Containment |  CDA RegionOfInterest CDA RegionOfInterest | 2005‑09‑07 | | 1.3.6.1.4.1.19376.1.5.3.1.4.2 | Containment |  IHE Comment Entry (2014) IHE Comment Entry (2014) | 2013‑12‑20 | | 1.3.6.1.4.1.19376.1.8.1.4.9 | Containment |  AP Observation Entry (2.0) AP Observation Entry (2.0) | 2014‑05‑14 17:09:54 | | 1.3.6.1.4.1.19376.1.3.1.6 | Containment |  Laboratory Observation (2017) Laboratory Observation (2017) | 2008‑08‑08 | | 2.16.840.1.113883.10.12.324 | Containment |  CDA Reference CDA Reference | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.305 (2005‑09‑07) |
|---|
| Example | | generated example | <organizer classCode="BATTERY" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.8.1.3.6"/> <id root="1.2.3.999" extension="--example only--"/> <code code="75326-9" codeSystem="2.16.840.1.113883.6.1" displayName="Problem">
<qualifier>
<name code="116677004" codeSystem="2.16.840.1.113883.6.96" displayName=" Associated topography (attribute)"/> <value nullFlavor="cs" code="all children of "362981000"" codeSystem="2.16.840.1.113883.6.96" displayName="all values from 91723000 (Anatomical structure)"/> </qualifier> </code> <component typeCode="COMP">
<observation>
<code code="75326-9" codeSystem="2.16.840.1.113883.6.1" displayName="Problem">
<value code="115597007" displayName="Description of specimen character (procedure)" codeSystem="2.16.840.1.113883.6.96"/> </code> </observation> </component> <statusCode code="completed"/> <effectiveTime>
<low value="20170131165954"/> </effectiveTime> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant> <participant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.7 'Additional participant in an entry APSR (Body)' (2016-07-12T18:44:06) -->
</participant> <performer>
<!-- template 1.3.6.1.4.1.19376.1.3.3.1.7 'Laboratory Performer' (2008-08-08T00:00:00) -->
</performer> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 0..* R -->
<component typeCode="COMP" contextConductionInd="true">
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.8.1.4.10 'Embedded Image IHE' (2016-06-22T16:08:01) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 2.16.840.1.113883.10.12.307 'CDA RegionOfInterest' (2005-09-07T00:00:00) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.5.3.1.4.2 'IHE Comment Entry' (2013-12-20T00:00:00) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.8.1.4.9 'AP Observation Entry' (2014-05-14T17:09:54) -->
</component> <component typeCode="COMP" contextConductionInd="true">
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.3.1.6 'Laboratory Observation' (2008-08-08T00:00:00) -->
</component> <reference>
<!-- template 2.16.840.1.113883.10.12.324 'CDA Reference' (2005-09-07T00:00:00) -->
</reference></organizer> |
|
| Example | | example for use case #1, problem: invasive breast cancer | <organizer classCode="BATTERY" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.8.1.3.6"/> <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_Problem1"/> <code code="75326-9" codeSystem="2.16.840.1.113883.6.1" displayName="Problem"/> <statusCode code="completed"/> <effectiveTime>
<low value="201001041605-0500"/> </effectiveTime> <!--The problem is expressed by the result of the observation having the same code as organizer/code-->
<component>
<observation classCode="OBS" moodCode="EVN">
<code code="75326-9" codeSystem="2.16.840.1.113883.6.1" displayName="Problem"/> <value code="C50.9" codeSystem="2.16.840.1.113883.6.3" displayName="Malignant neoplasm of breast, unspecified" codeSystemDisplayName="ICD-10"/> </observation> </component> <component typeCode="COMP" contextConductionInd="true">
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.8.1.4.9 'AP Observation Entry' (2014-05-14T17:09:54) -->
</component> <reference>
<!-- template 2.16.840.1.113883.10.12.324 'CDA Reference' (2005-09-07T00:00:00) -->
</reference></organizer> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … * | R | Problem organizer groups the battery of observations performed to investigate on a single problem identified on a specimen or a group of specimens. | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | @classCode
|
| cs | 1 … 1 | F | BATTERY |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | hl7:templateId
|
| II | 1 … 1 | M | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.8.1.3.6 |  | hl7:id
|
| II | 0 … * | R | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:code
|
| CD (required) | 0 … 1 | R | Problem
| PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | @code
|
| CONF | 0 … 1 | F | 75326-9 |  |  | @codeSystem
|
| 0 … 1 | F | 2.16.840.1.113883.6.1 (Logical Observation Identifier Names and Codes) |  |  | @codeSystemName
|
| 0 … 1 | F | LOINC |  |  | @displayName
|
| 0 … 1 | F | Problem |  | hl7:component
|
| | 0 … 1 | R | Coding of the Problem reported | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | hl7:observation
|
| | 0 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | CD (extensible) | 1 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | CONF | 0 … 1 | F | 75326-9 | | 0 … 1 | F | 2.16.840.1.113883.6.1 (Logical Observation Identifier Names and Codes) | | 0 … 1 | F | LOINC | | 0 … 1 | F | Problem | | CR | 0 … * | R | Optional qualifier for separate problems with identical problem type in one specimen, e.g. a urothelial bladder cancer Invasive cancer) and a prostate cancer (invasive cancer) in one cystoprostatectomy specimen. The anatomic structure is able to differentiate.
| PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | | Example | Problem in urinary bladder in cystoprostatectomy specimen <qualifier>
<name code="116677004" codeSystem="2.16.840.1.113883.6.96" displayName="Associated topography (attribute)" codeSystemName="SNOMED-CT"/> <value code="302512001" codeSystem="2.16.840.1.113883.6.96" displayName=" Entire urinary bladder (body structure)" codeSystemName="SNOMED-CT"/></qualifier> | | CV | 0 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | cs | 1 … 1 | F | 116677004 | | oid | 1 … 1 | F | 2.16.840.1.113883.6.96 | | st | 1 … 1 | F | Associated topography (attribute) | | CD (extensible) | 0 … * | R | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | CONF | 0 … 1 | F | << 91723000 | | 0 … 1 | F | 2.16.840.1.113883.6.96 (SNOMED Clinical Terms) | | 0 … 1 | F | SNOMED-CT | | 0 … 1 | F | all values from Anatomical structure | | CE (extensible) | 0 … 1 | R | Code of the problem reported | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | | CONF | | @code shall be "any_code" | | @codeSystem shall be "2.16.840.1.113883.6.3" | | @codeSystemName shall be "any_code" | | @displayName shall be "ICD-10" | | or | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.7 Problem Type (2016‑07‑11 09:52:19) |
|  | hl7:statusCode
|
| CS (required) | 1 … 1 | M | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer | | | Constraint | The status of the organizer is
completed
if the battery of observations for this problem has been performed. It is
aborted
in case some of the intended observations could not be achieve and have been aborted, and appear as such below the organizer.
| | | CONF | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.2 (DYNAMIC) | Error: Cannot find value set "1.3.6.1.4.1.19376.1.3.11.2" |
|
|  | hl7:effectiveTime
|
| IVL_TS | 1 … 1 | M | Organizer time.
The period of time (possibly reduced to a point in time) during which the information was collected and assembled.
| PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:author
|
| | 0 … * | C | Author
Source: PaLM Suppl. APSR 2.0-3: 6.3.6.2
Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| | | | Constraint | The author SHALL be present if it is different from the main author of the report. For instance the author could be the ordering physician contributing to the Clinical Information section, inside a report authored by the pathologist.
|  | hl7:informant
|
| | 0 … * | R | Informant
The informant is an actor (person and organization) who provided some of the clinical information carried by this organizer.
Source: PaLM Suppl. APSR 2.0-3:
6.3.6.5
Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| |  | hl7:participant
|
| | 0 … * | R | Transcriptionist, Device, Responsible, Validator
Source: PaLM Suppl. APSR 2.0-3:
6.3.6.6
Contains 1.3.6.1.4.1.19376.1.8.1.4.7 Additional participant in an entry APSR (Body) (2016‑07‑12 18:44:06) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.7']] |
| | | | Constraint | participationType "ENT", "DEV", "RESP", "AUTHEN"
|  | hl7:performer
|
| | 0 … 1 | C | Performing lab.
The performing laboratory is present at this level only if this particular problem on this particular specimen was investigated by a (subcontractor) distinct laboratory from the one issuing the consolidated report
Source:
PaLM TF-3: 6.3.2.20
Contains 1.3.6.1.4.1.19376.1.3.3.1.7 Laboratory Performer (2008‑08‑08) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.7']] |
| | | Included | 0 … * | R | from 1.3.6.1.4.1.19376.1.3.10.9.1 X Specimen Identified (2014‑12‑22 15:54:30)
Specimen identification outside an AP observation for differentiating or restituting the problem type
Source: PaLM Suppl. APSR 2.0-3: 6.3.6.9
|  | hl7:specimen
|
| | 0 … * | R | Required in XD-LAB when more than one specimen is documented at this level. Always required in APSR.
| PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  |  | @typeCode
|
| cs | 0 … 1 | F | SPC |  |  | hl7:specimenRole
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified | | cs | 0 … 1 | F | SPEC | | II | 1 … 1 | R | Specimen ID, coming from the LIS | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  | hl7:component
|
| | 0 … * | R | Embedded image
This organizer may carry an image focusing on the problem, either directly embedded or encapsulated within a region of interest.
Contains 1.3.6.1.4.1.19376.1.8.1.4.10 Embedded Image IHE (2016‑06‑22 16:08:01) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [hl7:observationMedia [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.10']]] |
| |  |  | @typeCode
|
| cs | 0 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true | | | Constraint | [@classCode="ROIOVL" and moodCode="EVN"] |  |  | hl7:sequenceNumber
|
| INT | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | hl7:seperatableInd
|
| BL | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:component
|
| | 0 … * | R | Region of interest
This organizer may carry an image focusing on the problem, either directly embedded or encapsulated within a region of interest.
In the latter case by an entryRelationship @typeCode="SUBJ"
Contains 2.16.840.1.113883.10.12.307 CDA RegionOfInterest (2005‑09‑07) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [@typeCode='COMP'] [hl7:regionOfInterest [hl7:templateId [@root='2.16.840.1.113883.10.12.307']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true | | | Constraint | [@classCode="ROIOVL" and moodCode="EVN"] |  |  | hl7:sequenceNumber
|
| INT | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | hl7:seperatableInd
|
| BL | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:component
|
| | 0 … * | R | General comment on this particular problem
Source: PCC TF-2: 6.3.4.6
Contains 1.3.6.1.4.1.19376.1.5.3.1.4.2 IHE Comment Entry (2013‑12‑20) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [@typeCode='COMP'] [hl7:act [hl7:templateId [@root='2.16.840.1.113883.10.20.1.40'] and hl7:templateId [@root='1.3.6.1.4.1.19376.1.5.3.1.4.2']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true |  |  | hl7:sequenceNumber
|
| INT | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | hl7:seperatableInd
|
| BL | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:component
|
| | 0 … * | R | Any kind of AP observation, for complex observations, e.g. for TNM, ICD-O-3, or Assessment scales, the appropriate templates may be used.
Source: PaLM Suppl. APSR 2.0-3: 6.3.6.7
Contains 1.3.6.1.4.1.19376.1.8.1.4.9 AP Observation Entry (2014‑05‑14 17:09:54) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [@typeCode='COMP'] [hl7:observation [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.9']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true |  |  | hl7:sequenceNumber
|
| INT | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | hl7:seperatableInd
|
| BL | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:component
|
| | 0 … * | C | Laboratory observation
Contains 1.3.6.1.4.1.19376.1.3.1.6 Laboratory Observation (2008‑08‑08) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | where [@typeCode='COMP'] [hl7:observation [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.1.6']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true |  |  | hl7:sequenceNumber
|
| INT | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  |  | hl7:seperatableInd
|
| BL | 0 … 1 | | | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |  | hl7:reference
|
| | 0 … * | R | Reference to an external document, or to external organizers, or to external observations, e.g. concerning molecular pathology or genetic testing report
Contains 2.16.840.1.113883.10.12.324 CDA Reference (2005‑09‑07) | PaLM Suppl. 2.0‑3: 6.3.5.2 Problem organizer |
|
6.3.5.3 Specimen Procedure Step <entry> - 1.3.6.1.4.1.19376.1.3.10.4.1
6.3.5.3.1 Definition and purpose
The Specimen Procedure Step <entry> Content Module contains in machine-readable form all the data concerning one specimen or a group of specimens together with its container(s). It is usable within a Procedure Steps <section> Content Module only.
This Content Module structures the machine-readable data, which characterize the specimens and the procedures of their collection and processing (identifiers and types, status, time intervals, performers, approach site, target site, etc.).
By referencing the child procedure ID via an entryRelationship the entry is used in an iterative manner for each single step of specimen collection and processing, which results in derived (child) specimen.
The resulting specimen is the participant in the procedure according to the CDA-RIM (fig. 6.2.5.3.2-1) with the typeCode="PRD", and the ParticipantRole.classCode="SPEC". The specimen ID is the ParticipantRole.ID.
Additives to the specimen (e.g. fixatives) MAY be additional participants with the typeCode="CSM" and the ParticipantRole.classCode="ADTV".
Each specimen is put in or on an appropriate container. The container is described through one or more EntryRelationship(s) to one or more Supply whose product is the container and possible container components (see this volume, 6.3.6.4).
The specimen reception in laboratory may be expressed through another EntryRelationship of typeCode "COMP", introducing the "specimen received" Act.
In case the procedure step produces a child specimen, the parent specimen is the specimen on which the procedure is performed. Thus the parent specimen is represented by a Specimen participation to the procedure. In this case, Specimen.SpecimenRole.Id is the id of the parent specimen. These parent specimens might be multiple, for instance in the case of TMA.Therefore, cardinalities of Specimen are [0..*]
Specimen and their containers carry IDs as well as further attributes, which are defined in the HL7 Specimen DAM [9].
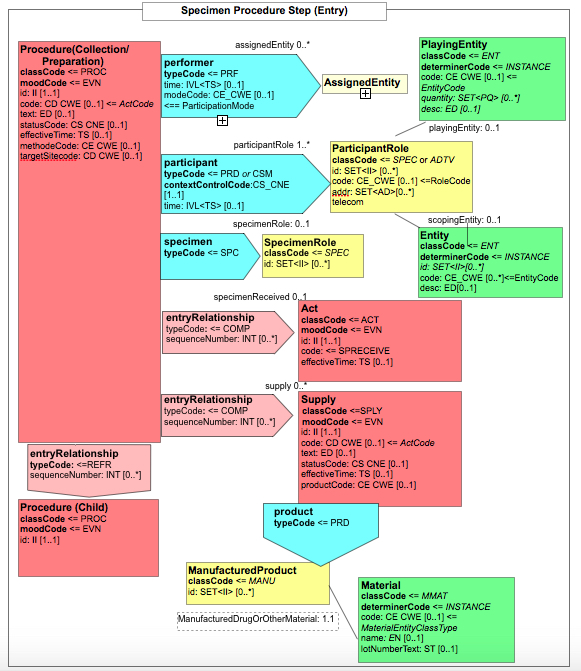
Figure 6.3.5.3.1-1 CDA-R-MIM of the Specimen Procedure Steps <entry>
According to the CDA RIM in the Content Module further supporting Content Modules for Participant, and Playing Entity are needed, which have the OIDs 1.3.6.1.4.1.19376.1.3.10.9.44, 1.3.6.1.4.1.19376.1.3.10.9.21).
The Code systems and Value sets MAY be organized organ-specific.
6.3.5.3.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.3.10.4.1 | Effective Date | valid from 2016‑07‑08 13:20:59There are versions of templates with this id:- SpecimenProcedureStep as of 2016‑07‑08 13:20:59
- SpecimenProcedureStep as of 2014‑07‑29 16:02:02
|
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | SpecimenProcedureStep | Display Name | Specimen Procedure Step |
|---|
| Description | Template CDA Procedure (prototype, directly derived from POCD_RM000040 MIF)
for Specimen procedure step in APSR2.
This entry collects all the information of source, method and result of a specimen procedure from the collection out of the patient through the final product of a stained histologic section or cytologic smear on a glass slide, or a WSI of such a slide. It reflects the workflow and the probe tracking of a pathology laboratory.
|
|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.4.1 |
|---|
| Label | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps
|
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 4 templates, Uses 8 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.1 | Containment |  Specimen Procedure Step (2.0) Specimen Procedure Step (2.0) | 2016‑07‑08 13:20:59 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.8.1.2.6 |  |  Procedure Steps Section (2.0) Procedure Steps Section (2.0) | 2014‑05‑13 19:33:12 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.3.10.9.44 | Containment |  CDA Participant (Body) Procedure Steps APSR2 (2.0) CDA Participant (Body) Procedure Steps APSR2 (2.0) | 2014‑10‑26 17:06:28 | | 1.3.6.1.4.1.19376.1.3.10.9.22 | Containment |  CDA Supply Container APSR2 (2.0) CDA Supply Container APSR2 (2.0) | 2016‑07‑29 12:09:33 | | 1.3.6.1.4.1.19376.1.3.1.3 | Containment |  Specimen Received (2017) Specimen Received (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.3.10.4.1 | Containment |  Specimen Procedure Step (2.0) Specimen Procedure Step (2.0) | 2016‑07‑08 13:20:59 | | 2.16.840.1.113883.10.12.324 | Containment |  CDA Reference CDA Reference | 2005‑09‑07 |
|
|
|---|
| Relationship | Adaptation: template 2.16.840.1.113883.10.12.306 (2005‑09‑07) |
|---|
| Example | | generated example | <procedure negationInd="false" classCode="PROC" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.10.4.1"/> <id root="1.2.3.999" extension="--example only--"/> <code code="117617002" displayName="Immunohistochemistry procedure (procedure)" codeSystem="2.16.840.1.113883.5.96">
<qualifier>
<name code="246094008 or 246355002" codeSystem="2.16.840.1.113883.6.96" displayName="Component investigated (attribute) or Supporting structure (attribute)"/> <value code="23307004" codeSystem="2.16.840.1.113883.6.96" displayName="Estrogen receptor (substance)"/> </qualifier> </code> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20161110104817"/> </effectiveTime> <targetSiteCode code="441652008" codeSystem="2.16.840.1.113883.6.96" displayName="Formalin-fixed paraffin-embedded tissue specimen (specimen)"/> <specimen typeCode="SPC">
<specimenRole classCode="SPEC">
<id/> </specimenRole> </specimen> <performer>
<!-- template 1.3.6.1.4.1.19376.1.3.3.1.7 'Laboratory Performer' (2008-08-08T00:00:00) -->
</performer> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <participant>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.44 'CDA Participant (Body) Procedure Steps APSR2' (2014-10-26T17:06:28) -->
</participant> <entryRelationship negationInd="false" typeCode="COMP" inversionInd="false" contextConductionInd="true">
<seperatableInd value="false"/> <sequenceNumber value="1"/> <!-- template 1.3.6.1.4.1.19376.1.3.10.9.22 'CDA Supply Container APSR2' (2016-07-29T12:09:33) -->
</entryRelationship> <entryRelationship negationInd="false" typeCode="COMP" inversionInd="false" contextConductionInd="true">
<seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.3.1.3 'Specimen Received' (2008-08-08T00:00:00) -->
</entryRelationship> <entryRelationship negationInd="false" typeCode="REFR" inversionInd="false" contextConductionInd="true">
<seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.3.10.4.1 'Specimen Procedure Step' (2016-07-08T13:20:59) -->
</entryRelationship> <reference>
<!-- template 2.16.840.1.113883.10.12.324 'CDA Reference' (2005-09-07T00:00:00) -->
</reference></procedure> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … * | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | @classCode
|
| cs | 1 … 1 | F | PROC |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | hl7:templateId
|
| II | 1 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.10.4.1 |  | hl7:id
|
| II | 0 … * | R | Procedure ID, coming from the LIS.
May be the target (referenced) ID for the child procedure. | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | hl7:code
|
| CD (extensible) | 1 … 1 | M | Coded procedure | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | | Constraint | for specimen collection procedure codes for targetSite or SNOMED CT have to be used, for specimen processing (down step the procedures) SNOMED CT is preferred, DICOM could be used if available
| | | CONF | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.10 Specimen Collection and Processing (2016‑05‑31 15:09:59) |
| | | Example | for use case #1, specimen collection <code code="122548005" codeSystem="2.16.840.1.113883.6.96" displayName="Biopsy of breast (procedure)" codeSystemName="SNOMED-CT"/> | | | Example | for use case #1, grossing & embedding <code code="40923002" codeSystem="2.16.840.1.113883.6.96" displayName="Tissue processing technique, routine, embed, cut and stain, per surgical specimen (procedure)"/> | | | Example | for use case #1, sectioning & immunostaining <code code="117617002" codeSystem="2.16.840.1.113883.6.96" displayName="Immunohistochemistry procedure (procedure)"/> |  |  | hl7:qualifier
|
| CR (extensible) | 0 … * | C | Qualifier for staining or other procedures which don´t have values in the vocabulary used.
| PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | | Constraint | Used only in staining and other procedures without values in SCT or LOINC or DICOM vocabulary, e.g. immune stains
| | | Example | for use case #1, grossing & embedding <qualifier>
<name code="246355002" displayName="Supporting structure (attribute)" codeSystem="2.16.840.1.113883.6.96"/> <value code="702941008" displayName="Paraffin embedding (qualifier value)" codeSystem="2.16.840.1.113883.6.96" codeSystemName="SNOMED-CT"/></qualifier> | | | Example | for use case #1, Estrogen receptor staining <qualifier>
<name code="246094008" displayName="Component investigated (attribute)" codeSystem="2.16.840.1.113883.6.96"/> <value code="23307004" displayName="Estrogen receptor (substance)" codeSystem="2.16.840.1.113883.6.96"/></qualifier> | | | Example | for use case #1, ISH Her-2-neu staining <qualifier>
<name code="246094008" displayName="Component investigated (attribute)" codeSystem="2.16.840.1.113883.6.96"/> <value code="164870" displayName="ONCOGENE ERBB2" codeSystem="2.16.840.1.113883.6.174" codeSystemName="OMIM"/></qualifier> | | CV (extensible) | 1 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | cs | 0 … 1 | F | 246094008_or_246355002 | | oid | 0 … 1 | F | 2.16.840.1.113883.6.96 | | st | 0 … 1 | F | Component investigated (attribute) or Supporting structure (attribute) | | CD (extensible) | 1 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | cs | 1 … 1 | R | | | | CONF | | @code shall be "<<105590001 | Substance (substance) |" | | @codeSystem shall be "2.16.840.1.113883.6.96" | | @codeSystemName shall be "SNOMED-CT" | | @displayName shall be "values from substance axis, e.g. Estrogen receptor" | | or | | @code shall be "values from G axis" | | @codeSystem shall be "2.16.840.1.113883.6.174" | | @codeSystemName shall be "OMIM" | | @displayName shall be "values from G axis, e.g. erb-b2" | | or | | @code shall be "<<272394005 | Technique (qualifier value) |" | | @codeSystem shall be "2.16.840.1.113883.6.96" | | @codeSystemName shall be "SNOMED-CT" | | @displayName shall be "values from technique axis, e.g. Paraffin embedded (qualifier)" |
|  | hl7:text
|
| ED | 0 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | | Example | specimen collection procedure <text>ULTRASOUND GUIDED NEEDLE CORE BIOPSY</text> | | | Example | specimen processing procedure <text>CUTTING A SLIDE FROM A PARAFFIN BLOCK</text> |  | hl7:statusCode
|
| CS (required) | 0 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | | CONF | | The value of @code shall be drawn from value set 2.16.840.1.113883.1.11.15933 ActStatus (DYNAMIC) |
|  | hl7:effectiveTime
|
| IVL_TS | 0 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | hl7:approachSiteCode
|
| CD (extensible) | 0 … 1 | R | Approach site in specimen collection | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  |  | @code
|
| CONF | 0 … 1 | F | any_code |  |  | @codeSystem
|
| 0 … 1 | F | 2.16.840.1.113883.5.1052 (Act Site) |  |  | @codeSystemName
|
| 0 … 1 | F | any_code |  |  | @displayName
|
| 0 … 1 | F | ActSite |  | hl7:targetSiteCode
|
| CD (extensible) | 0 … 1 | R | Target site in body or relative parent relationship in case of specimen processing | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | | CONF | | @code shall be "<<123037004 | Body structure (body structure) |" | | @codeSystem shall be "2.16.840.1.113883.6.96" | | @codeSystemName shall be "SCT" | | @displayName shall be "all children of Body structure (body structure)" | | or | | @code shall be "any_code" | | @codeSystem shall be "2.16.840.1.113883.5.1052" | | @codeSystemName shall be "ActSite" | | @displayName shall be "any_code" | | or | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.9 Procedure Site (2016‑06‑01 18:46:15) |
| | | Example | for use case #1, specimen collection <targetSiteCode code="110497008" displayName="Lower outer quadrant of right breast (body structure)" codeSystem="2.16.840.1.113883.6.96"/> | | | Example | for use case #1, grossing & embedding <targetSiteCode code="309050000" displayName="Body substance sample" codeSystem="2.16.840.1.113883.6.96"/> |  |  | hl7:qualifier
|
| CR (extensible) | 0 … * | C | optional qualifiers for specifying body structures or / and laterality | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | | Example | for use case #1, (optional) specification of body structure for specimen collection <qualifier>
<name code="118169006" codeSystem="2.16.840.1.113883.6.96" displayName="Specimen source topography (attribute)" codeSystemName="SNOMED-CT"/> <value code="255495000" codeSystem="2.16.840.1.113883.6.96" displayName="Right lower quadrant (qualifier value)" codeSystemName="SNOMED-CT"/></qualifier> | | CV (extensible) | 1 … 1 | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | st | 0 … 1 | F | Specimen source topography | | oid | 1 … 1 | F | 2.16.840.1.113883.6.96 | | cs | 1 … 1 | F | 118169006 | | CD (extensible) | 0 … * | R | | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps | | CONF | 0 … 1 | | @code shall be out of choice:- <<272099008
- Descriptor (qualifier value)
| | 0 … 1 | F | 2.16.840.1.113883.6.96 (SNOMED Clinical Terms) | | 0 … 1 | F | SCT | | 0 … 1 | F | all children of "Descriptor, qualifier values", e.g. right lower quadrantt |  | hl7:methodCode
|
| CD (extensible) | 0 … * | R | Identifies the technique used to perform a procedure
| PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  |  | @codeSystem
|
| cs | 1 … 1 | F | 2.16.840.1.113883.5.1065 | | Included | 0 … 1 | R | from 1.3.6.1.4.1.19376.1.3.10.9.1 X Specimen Identified (2014‑12‑22 15:54:30)
Parent specimen with parent ID | | | Example | for use case #1, parent specimen ID in grossing & embedding <specimen typeCode="SPC">
<specimenRole classCode="SPEC">
<!-- Parent specimen ID -->
<id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008"/> </specimenRole></specimen> |  | hl7:specimen
|
| | 0 … 1 | R | Required in XD-LAB when more than one specimen is documented at this level. Always required in APSR.
| PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  |  | @typeCode
|
| cs | 0 … 1 | F | SPC |  |  | hl7:specimenRole
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified | | cs | 0 … 1 | F | SPEC | | II | 1 … 1 | R | Specimen ID, coming from the LIS | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  | hl7:performer
|
| | 0 … 1 | R | Contains 1.3.6.1.4.1.19376.1.3.3.1.7 Laboratory Performer (2008‑08‑08) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.7']] |
| |  | hl7:author
|
| | 0 … * | C | Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| |  | hl7:participant
|
| | 1 … * | M | One or more specimen (with additives), which participates as product of the procedure
Contains 1.3.6.1.4.1.19376.1.3.10.9.44 CDA Participant (Body) Procedure Steps APSR2 (2014‑10‑26 17:06:28) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | where [not(@nullFlavor)] |
| |  | hl7:entryRelationship
|
| | 0 … * | R | Artificial material (manufactured product) which
containes
the specimen
Contains 1.3.6.1.4.1.19376.1.3.10.9.22 CDA Supply Container APSR2 (2016‑07‑29 12:09:33) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | where [@typeCode='COMP'] [hl7:supply [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.10.9.22']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true | | | Constraint | for manufactured product
classCode = MANU
for DrugOrOtherMaterial
classCode = MMAT
code from MaterialEntityClassType
|  | hl7:entryRelationship
|
| | 0 … 1 | R | Specimen arrival in Lab
Contains 1.3.6.1.4.1.19376.1.3.1.3 Specimen Received (2008‑08‑08) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | where [@typeCode='COMP'] [hl7:act [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.1.3']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true |  | hl7:entryRelationship
|
| | 0 … * | R | Referencing mechanism to the ID of the child procedures, giving a lis of child procedure IDs
Contains 1.3.6.1.4.1.19376.1.3.10.4.1 Specimen Procedure Step (2016‑07‑08 13:20:59) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |  | where [@typeCode='REFR'] [hl7:procedure [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.10.4.1']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | REFR |  |  | @contextConductionInd
|
| bl | 0 … 1 | F | true | | | Constraint | contains only ID of the child procedure, which is fully described in a dedicated entry further down in the Procedure step section. |  | hl7:reference
|
| | 0 … * | R | Reference to an external document, or to external procedures, e.g. concerning molecular pathology or genetic testing report
Contains 2.16.840.1.113883.10.12.324 CDA Reference (2005‑09‑07) | PaLM Suppl. 2.0‑3: 6.3.5.3 Specimen procedure steps |
|
Supportive template CDA Participant (Body) Procedure Steps APSR2 - 1.3.6.1.4.1.19376.1.3.10.9.44
Supportive template CDA PlayingEntitySpecimen APSR2 - 1.3.6.1.4.1.19376.1.3.10.9.21
6.3.5.4 Update Information Organizer <entry> - 1.3.6.1.4.1.19376.1.3.10.4.5
6.3.5.4.1 Definition and purpose
This content module is usable in the Diagnostic conclusion section only.
It is used in the situation where a new version of a report is replacing the previous one, as to indicate what sections and/or entries have been changed in comparison with the immediate previous version of the report.
6.3.5.4.2 Specification and Example
6.3.6 CDA R2 Child Element Content Modules
This section specifies the Content Modules designed for child elements. A child element is a child of the CDA header or a child of a <section>, or an element nested at various depths below an <entry>, or an element appearing at some combination of these locations.
6.3.6.1.1 Definition and purpose
This Content Module is usable only in the CDA header.
This Content Module is used only in the situation where the specimen was not collected by the ordering physician. (See use cases in volume 1)
6.3.6.1.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.8.1.4.1 | Effective Date | valid from 2016‑06‑13 14:21:13 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | CDAParticipantSpecimenCollector | Display Name | CDA Participant Specimen Collector |
|---|
| Description | Template CDA participant (prototype, directly derived from POCD_RM000040 MIF)
This Content Module is usable only in the CDA header.
This Content Module is used only in the situation where the specimen was not collected by the ordering physician.
|
|
| Label | PaLM Suppl. APSR 2.0‑3:6.3.6.1 Specimen collector
|
|---|
| Classification | CDA Header Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 1 template, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.1.1 | Include |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | Uses | as | Name | Version |
|---|
| 2.16.840.1.113883.10.12.152 | Containment |  CDA Person CDA Person | 2005‑09‑07 | | 2.16.840.1.113883.10.12.151 | Containment |  CDA Organization CDA Organization | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.108 (2005‑09‑07) |
|---|
| Example | | generated example | <participant typeCode="DIST" contextControlCode="OP">
<templateID root="1.3.6.1.4.1.19376.1.8.1.4.1"/> <time>
<low value="20170127121035"/> </time> <associatedEntity classCode="PROV">
<id root="1.2.3.999" extension="--example only--"/> <code code="--code--" codeSystem="2.16.840.1.113883.5.111"/> <addr>addr</addr> <telecom/> <associatedPerson>
<!-- template 2.16.840.1.113883.10.12.152 'CDA Person' (dynamic) -->
</associatedPerson> <scopingOrganization>
<!-- template 2.16.840.1.113883.10.12.151 'CDA Organization' (dynamic) -->
</scopingOrganization> </associatedEntity></participant> |
|
|
6.3.6.2 Author – 1.3.6.1.4.1.19376.1.8.1.4.2
6.3.6.2.1 Definition and purpose
This Content Module is usable in the CDA header, in a <section> and at various depths of an <entry>.
It describes an author having contributed to the document wholly or to a portion of it (e.g., a section, an observation, a group of observations).
A given document or any delimited portion of it may have more than one author.
An author MAY be a person or a device (manufactured device or software system). In both cases the scoping organization MAY be described.
6.3.6.2.2 Specification and Example
6.3.6.3 Content Validator – 1.3.6.1.4.1.19376.1.8.1.4.3
6.3.6.3.1 Definition and purpose
This Content Module is usable only in the CDA header.
It describes a pathologist having verified the content of the report, using the element authenticator. This element authenticator is used when the pathologist having verified the content of the report is distinct from the pathologist assuming the legal responsibility for this report, described in the legalAuthenticator element.
The report MAY have zero or more Content Validators.
6.3.6.3.2 Specification and Example
6.3.6.4 Specimen Container in Specimen Procedure Step - 1.3.6.1.4.1.19376.1.3.10.9.22
6.3.6.4.1 Definition and purpose
This Supporting Content Module is usable in the Specimen Procedure Steps <entry> (see 6.2.6.4) and is repeatable within one procedure step for describing container components, too. It describes the properties of containers, in or on which the specimens are processed. The relations between container and specimen are described in the HL7 Specimen DAM.
HL7 DAM Specimen Release 1.
For Anatomic Pathology the CDA Content Modules "Material" and "ManufacturedMaterial" were specialized to Supporting Templates CDA ManufacturedProduct APSR2 (1.3.6.1.4.1.19376.1.3.10.9.24) and CDA Material Container APSR2 (1.3.6.1.4.1.19376.1.3.10.9.23).
6.3.6.4.2 Specification and Example
Supportive Template CDA ManufacturedProduct APSR2 - 1.3.6.1.4.1.19376.1.3.10.9.24
| Id | 1.3.6.1.4.1.19376.1.3.10.9.24 | Effective Date | valid from 2016‑09‑09 12:37:49 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | CDAManufacturedProductAPSR2 | Display Name | CDA ManufacturedProduct APSR2 |
|---|
| Description | Template CDA ManufacturedProduct (prototype, directly derived from POCD_RM000040 MIF)
Supporting Content Module for CDA Supply Container APSR2, bridging the gap between Product and Container |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 5 templates, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.22 | Containment |  CDA Supply Container APSR2 (2.0) CDA Supply Container APSR2 (2.0) | 2016‑07‑29 12:09:33 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (2.0) Specimen Procedure Step (2.0) | 2016‑07‑08 13:20:59 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.8.1.2.6 |  |  Procedure Steps Section (2.0) Procedure Steps Section (2.0) | 2014‑05‑13 19:33:12 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.23 | Include |  CDA Material Container APSR2 CDA Material Container APSR2 | 2016‑09‑08 12:20:45 | | 2.16.840.1.113883.10.12.151 | Containment |  CDA Organization CDA Organization | 2005‑09‑07 |
|
|
|---|
| Relationship | Adaptation: template 2.16.840.1.113883.10.12.312 (2005‑09‑07) |
|---|
| Example | | Example | <manufacturedProduct classCode="MANU">
<id root="1.2.3.999" extension="--example only--"/> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.23 'CDA Material Container APSR2' (2016-09-08T12:20:45) 0..1 R -->
<manufacturerOrganization>
<!-- template 2.16.840.1.113883.10.12.151 'CDA Organization' (2005-09-07T00:00:00) -->
</manufacturerOrganization></manufacturedProduct> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … 1 | R | | (CDA...SR2) |  | @classCode
|
| cs | 1 … 1 | F | MANU |  | hl7:id
|
| II | 0 … * | R | | (CDA...SR2) | | Included | 1 … 1 | M | from 1.3.6.1.4.1.19376.1.3.10.9.23 CDA Material Container APSR2 (2016‑09‑08 12:20:45)
Material the specimen container consists of, e.g. Tissue cassette or Paraffin wax for a Paraffin Block, etc. |  | hl7:manufacturedMaterial
|
| | 1 … 1 | M | | (CDA...SR2) |  |  | @classCode
|
| cs | 1 … 1 | F | MMAT | | | class code "MMAT" for container |  |  | @determinerCode
|
| cs | 0 … 1 | F | KIND |  |  | hl7:code
|
| CE (extensible) | 0 … * | R | | (CDA...SR2) | | | CONF | | The value of @code should be drawn from value set 2.16.840.1.113883.1.11.16041 MaterialEntityClassType (2014‑03‑26) | | or | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.8 ContainerEntityClassTypePaLM (2016‑09‑08 11:55:11) |
|  |  | hl7:name
|
| EN | 0 … 1 | R | | (CDA...SR2) |  |  | hl7:lotNumberText
|
| ST | 0 … 1 | | | (CDA...SR2) |  | hl7:manufacturerOrganization
|
| | 0 … 1 | R | Contains 2.16.840.1.113883.10.12.151 CDA Organization (2005‑09‑07) | (CDA...SR2) |
|
Supportive Template CDA Material Container APSR2 - 1.3.6.1.4.1.19376.1.3.10.9.23
| Id | 1.3.6.1.4.1.19376.1.3.10.9.23 | Effective Date | valid from 2016‑09‑08 12:20:45 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | CDAMaterialContainerAPSR2 | Display Name | CDA Material Container APSR2 |
|---|
| Description | Template CDA Material (prototype, directly derived from POCD_RM000040 MIF)
Entry describing the properties of a container in PaLM / PAT |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 6 templates, Uses 0 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.24 | Include |  CDA ManufacturedProduct APSR2 CDA ManufacturedProduct APSR2 | 2016‑09‑09 12:37:49 | | 1.3.6.1.4.1.19376.1.3.10.9.22 |  |  CDA Supply Container APSR2 (2.0) CDA Supply Container APSR2 (2.0) | 2016‑07‑29 12:09:33 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (2.0) Specimen Procedure Step (2.0) | 2016‑07‑08 13:20:59 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.8.1.2.6 |  |  Procedure Steps Section (2.0) Procedure Steps Section (2.0) | 2014‑05‑13 19:33:12 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 |
|
|
|---|
| Relationship | Adaptation: template 2.16.840.1.113883.10.12.311 (2005‑09‑07) |
|---|
| Example | | generated example | <manufacturedMaterial classCode="MMAT" determinerCode="KIND">
<code code="PKG" displayName="Package" codeSystem="2.16.840.1.113883.5.1060"/> <name>name</name> <lotNumberText/></manufacturedMaterial> |
|
| Example | | example for use case '1, specimen collection | <manufacturedMaterial classCode="MMAT" determinerCode="KIND">
<code code="TUBE" displayName="tube" codeSystem="2.16.840.1.113883.1.11.16041"/> <name>Plastic tube</name></manufacturedMaterial> |
|
| Example | | example for use case '1, embedding | <manufacturedMaterial classCode="MMAT" determinerCode="KIND">
<code code="434464009" displayName="Tissue cassette (physical object)" codeSystem="2.16.840.1.113883.6.96"/> <name>Block</name></manufacturedMaterial> |
|
| Example | | example for use case '1, sectioning and staining | <manufacturedMaterial classCode="MMAT" determinerCode="KIND">
<code code="433466003" displayName="Microscope slide (physical object)" codeSystem="2.16.840.1.113883.6.96"/> <name>Slide</name></manufacturedMaterial> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
hl7:manufacturedMaterial
|
| | 0 … 1 | R | | (CDA...SR2) |  | @classCode
|
| cs | 1 … 1 | F | MMAT | | | class code "MMAT" for container |  | @determinerCode
|
| cs | 0 … 1 | F | KIND |  | hl7:code
|
| CE (extensible) | 0 … * | R | | (CDA...SR2) | | | CONF | | The value of @code should be drawn from value set 2.16.840.1.113883.1.11.16041 MaterialEntityClassType (2014‑03‑26) | | or | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.8 ContainerEntityClassTypePaLM (2016‑09‑08 11:55:11) |
|  | hl7:name
|
| EN | 0 … 1 | R | | (CDA...SR2) |  | hl7:lotNumberText
|
| ST | 0 … 1 | | | (CDA...SR2) |
|
6.3.6.5 Informant – 1.3.6.1.4.1.19376.1.8.1.4.6
6.3.6.5.1 Definition and purpose
This Content Module is usable in the CDA header, in a <section> and within an <entry>.
It describes a person having provided some piece of relevant information for the document.
A <ClinicalDocument> or a <section> or any kind of act below an <entry>, MAY have zero or more informant.
6.3.6.5.2 Specification and Example
6.3.6.6 Additional participant in an entry - 1.3.6.1.4.1.19376.1.8.1.4.7
6.3.6.6.1 Definition and purpose
This Content Module is usable only within an <entry> element.
Additional participants MAY take part in any organizer as well as in any observation of an APSR. These participants MAY be any of these 4:
- Validator: This is the same participation as Content Validator in the header of the report: a pathologist having verified the content (of this particular subset of results).
- Device: A device used to produce this particular subset of results.
- Responsible: The director of a laboratory (described in a performer element at the same level) who produced this particular subset of results.
- Transcriptionist: This is the same participation as dataEnterer in the header of the report: a staff who entered, possibly from dictation, this particular subset of results.
6.3.6.6.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.8.1.4.7 | Effective Date | valid from 2016‑07‑12 18:44:06 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | AdditionalParticipantAPSR | Display Name | Additional participant in an entry APSR (Body) |
|---|
| Description | Template CDA Participant (Body) (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Label | PaLM Suppl. APSR 2.0‑3: 6.3.6.6 Additional participant
|
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 18 templates, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | 1.3.6.1.4.1.19376.1.3.10.4.3 | Containment |  ICD-O-3 Typing and Grading Observation (2.0) ICD-O-3 Typing and Grading Observation (2.0) | 2015‑11‑29 17:03:18 | | 1.3.6.1.4.1.19376.1.3.10.4.4 | Containment |  Assessment Scales Observation for Scoring Systems APSR2 (2.0) Assessment Scales Observation for Scoring Systems APSR2 (2.0) | 2016‑10‑06 13:12:00 | | 1.3.6.1.4.1.19376.1.3.10.9.26 | Containment |  TNM N-Observation (2.0) TNM N-Observation (2.0) | 2014‑12‑02 16:34:50 | | 1.3.6.1.4.1.19376.1.3.10.9.27 | Containment |  TNM M-Observation (2.0) TNM M-Observation (2.0) | 2014‑12‑02 16:32:30 | | 1.3.6.1.4.1.19376.1.3.10.9.37 | Containment |  TNM Number of nodes (2.0) TNM Number of nodes (2.0) | 2014‑12‑02 16:48:36 | | 1.3.6.1.4.1.19376.1.3.10.9.43 | Containment |  Assessment Scoring Item APSR 2.0 (2.0) Assessment Scoring Item APSR 2.0 (2.0) | 2016‑10‑11 10:26:17 | | 1.3.6.1.4.1.19376.1.3.10.9.42 |  |  Assessment Scoring System APSR 2.0 (2.0) Assessment Scoring System APSR 2.0 (2.0) | 2016‑10‑10 11:33:36 | | 1.3.6.1.4.1.19376.1.8.1.3.6 | Containment |  Problem Organizer (2.0) Problem Organizer (2.0) | 2015‑08‑13 10:24:55 | | 1.3.6.1.4.1.19376.1.3.10.3.1 |  |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | 1.3.6.1.4.1.19376.1.8.1.2.1 |  |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 |  |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 |  |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 |  |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.8.1.2.5 |  |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | 1.3.6.1.4.1.19376.1.8.1.4.9 | Containment |  AP Observation Entry (2.0) AP Observation Entry (2.0) | 2014‑05‑14 17:09:54 | |  Circular reference found with 1.3.6.1.4.1.19376.1.8.1.4.9, please check Circular reference found with 1.3.6.1.4.1.19376.1.8.1.4.9, please check | | Uses | as | Name | Version |
|---|
| 2.16.840.1.113883.10.12.315 | Containment |  CDA Device CDA Device | 2005‑09‑07 | | 2.16.840.1.113883.10.12.313 | Containment |  CDA PlayingEntity CDA PlayingEntity | 2005‑09‑07 |
|
|
|---|
| Relationship | Adaptation: template 2.16.840.1.113883.10.12.321 (2005‑09‑07) |
|---|
| Example | | generated example | <templateId root="1.3.6.1.4.1.19376.1.8.1.4.7"/><time/><participantRole classCode="ROL">
<id root="1.2.3.999" extension="--example only--"/> <addr>addr</addr> <telecom value="tel:+1-12345678"/> <!-- choice: 0..1
element hl7:playingDevice containing template 2.16.840.1.113883.10.12.315 (dynamic)
element hl7:playingEntity containing template 2.16.840.1.113883.10.12.313 (dynamic)
-->
</participantRole> |
|
|
6.3.6.7 Anatomic Pathology Observation template – 1.3.6.1.4.1.19376.1.8.1.4.9
6.3.6.7.1 Definition and purpose
This Content Module is usable within a Problem Organizer.
The “AP Observation” generic template is usable for all AP observations, including those on findings from ancillary
techniques.
An AP Observation has a status and an effective time, MAY describe various participants (persons, devices, organizations), MAY have a number of additional properties (method,
interpretation, text), and MAY contain embedded images, comments, and sub-observations, which are also AP observations. For coding an AP observations LOINC codes SHOULD be used. For measurement results the UCUM system SHOULD be used.
Each kind of AP observation SHALL have one or more content modules "X specimen identified"
6.3.6.7.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.8.1.4.9 | Effective Date | valid from 2014‑05‑14 17:09:54 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | APObservationEntry | Display Name | AP Observation Entry |
|---|
| Description | This content module is usable within a Problem organizer.
The “AP Observation” generic template is usable for all AP observations, including ancillary techniques.
An AP observation has a status and an effective time, may describe various participants (persons, devices, organizations), may have a number of additional properties (method, interpretation, text), and may contain embedded images, comments, and sub-observations, which are also AP observations.
|
|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.8.1.4.9 |
|---|
| Label | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation
|
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 9 templates, Uses 10 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.3.6 | Containment |  Problem Organizer (2.0) Problem Organizer (2.0) | 2015‑08‑13 10:24:55 | | 1.3.6.1.4.1.19376.1.3.10.3.1 |  |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | 1.3.6.1.4.1.19376.1.8.1.2.1 |  |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 |  |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 |  |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 |  |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.8.1.2.5 |  |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | 1.3.6.1.4.1.19376.1.8.1.4.9 | Containment |  AP Observation Entry (2.0) AP Observation Entry (2.0) | 2014‑05‑14 17:09:54 | |  Circular reference found with 1.3.6.1.4.1.19376.1.8.1.4.9, please check Circular reference found with 1.3.6.1.4.1.19376.1.8.1.4.9, please check | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.8.1.4.9 | Containment |  AP Observation Entry (2.0) AP Observation Entry (2.0) | 2014‑05‑14 17:09:54 | | 1.3.6.1.4.1.19376.1.8.1.4.10 | Containment |  Embedded Image IHE Embedded Image IHE | 2016‑06‑22 16:08:01 | | 2.16.840.1.113883.10.12.307 | Containment |  CDA RegionOfInterest CDA RegionOfInterest | 2005‑09‑07 | | 1.3.6.1.4.1.19376.1.5.3.1.4.2 | Containment |  IHE Comment Entry (2014) IHE Comment Entry (2014) | 2013‑12‑20 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 2.16.840.1.113883.10.12.324 | Containment |  CDA Reference CDA Reference | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.303 (2005‑09‑07) |
|---|
| Example | | Observation example for use case #1, estrogen receptor | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.8.1.4.9"/> <id root="1.2.3.999" extension="--example only--"/> <code code="16112-5" codeSystem="2.16.840.1.113883.6.1" displayName="Estrogen receptor [Interpretation] in Tissue"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20161129162112"/> </effectiveTime> <value xsi:type="CD" code="416053008" displayName="Estrogen receptor positive tumor (disorder)" codeSystem="2.16.840.1.113883.6.96"/> <methodCode code="0107" displayName="Microscopy" codeSystem="2.16.840.1.113883.5.84"/> <performer>
<!-- template 1.3.6.1.4.1.19376.1.3.3.1.7 'CDA Performer IHE (Body)' (dynamic) -->
</performer> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author' (dynamic) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Body) APSR2' (dynamic) -->
</informant> <participant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.7 'Additional participant in an entry APSR (Body)' (dynamic) -->
</participant> <entryRelationship xsi:type="" typeCode="COMP">
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.9 'AP Observation Entry' (dynamic) -->
</entryRelationship> <entryRelationship xsi:type="" typeCode="COMP">
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.10 'Embedded Image IHE' (dynamic) -->
</entryRelationship> <entryRelationship xsi:type="" typeCode="COMP">
<!-- template 2.16.840.1.113883.10.12.307 'CDA RegionOfInterest' (dynamic) -->
</entryRelationship> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (dynamic) 0..* O -->
<reference>
<!-- template 2.16.840.1.113883.10.12.324 'CDA Reference' (dynamic) -->
</reference></observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … * | R | | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | @classCode
|
| cs | 1 … 1 | F | OBS |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | @negationInd
|
| bl | 0 … 1 | | |  | hl7:templateId
|
| II | 1 … 1 | M | | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.8.1.4.9 |  | hl7:id
|
| II | 0 … * | R | | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | hl7:code
|
| CE (extensible) | 1 … 1 | M | The Anatomic pathology observation code should be taken from a reference terminology, preferably the LOINC terminology. National terminologies may be used. | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  |  | @code
|
| CONF | 1 … 1 | F | each_code_for_observable_entities_or_a_specific_procedure |  |  | @codeSystem
|
| 1 … 1 | F | 2.16.840.1.113883.6.1 (Logical Observation Identifier Names and Codes) |  |  | @codeSystemName
|
| 1 … 1 | F | LOINC |  |  | @displayName
|
| 1 … 1 | F | see_above | | | Example | <code code="59847-4" codeSystem="2.16.840.1.113883.6.1" codeSystemName="LOINC" displayName="Histology and Behavior ICD-O-3"/> |  | hl7:text
|
| ED | 0 … 1 | R | Narrative text about the observation | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | hl7:statusCode
|
| CS (required) | 1 … 1 | M | The observation statusCode is “completed” if the observation was actually performed and has produced a result in the
value
element. In other cases the status of the intended observation is “aborted” and the result will never come. | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation | | | CONF | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.2 (DYNAMIC) | Error: Cannot find value set "1.3.6.1.4.1.19376.1.3.11.2" |
|
|  | hl7:effectiveTime
|
| IVL_TS | 1 … 1 | M | Date and time the observation has been made
| PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | hl7:value
|
| ANY | 0 … * | R | Observation values using the appropriate data type.
Numeric results use data type PQ, which includes the unit. The result is absent in case of ‘aborted’ observation.
| PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation | | | Constraint | The default data type, cardinalities and value set for the <value> element are dependant on the type of observation. | | | CONF | | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.11 AP Observation (2015‑11‑22 12:20:34) |
| | | Example | for observation on tumor type <value code="8500/3" codeSystem="2.16.840.1.113883.6.43.1" codeSystemName="ICD-O-3" displayName="Invasive carcinoma of the breast, no special type"/> |  | hl7:interpretationCode
|
| CE (extensible) | 0 … 1 | R | One or more codes interpreting the result, expressed with ObservationInterpretation vocabulary (e.g., H = high, L = low).
This element may be coded with other values than those of the ObservationInterpretation vocabulary, given that the coding strength is CWE (coded with exceptions).
| PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation | | | CONF | | The value of @code should be drawn from value set 2.16.840.1.113883.1.11.78 ObservationInterpretation (2014‑03‑26) |
|  | hl7:methodCode
|
| CE (extensible) | 0 … 1 | R | | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation | | | CONF | | The value of @code should be drawn from value set 2.16.840.1.113883.1.11.14079 ObservationMethod (2014‑03‑26) |
|  | hl7:performer
|
| II | 0 … 1 | C | Performing lab
Source: PaLM TF-3: 6.3.2.20
Contains 1.3.6.1.4.1.19376.1.3.3.1.7 Laboratory Performer (2008‑08‑08) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.7']] |
| | | | Constraint | The performing laboratory is present at this level only if this particular observation was performed by a (subcontractor) laboratory distinct from the one issuing the rest of the observations in this organizer.
|  | hl7:author
|
| II | 0 … * | C | Author
Source: PaLM TF Suppl. APSR 2.0-3:
6.3.6.2
Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| |  | hl7:informant
|
| II | 0 … * | R | Informant
Source: PaLM TF Suppl. APSR 2.0-3
: 6.3.6.5
Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| |  | hl7:participant
|
| II | 0 … * | R | Additional participants
Source: PaLM TF Suppl. APSR 2.0-3
: 6.3.6.6
Contains 1.3.6.1.4.1.19376.1.8.1.4.7 Additional participant in an entry APSR (Body) (2016‑07‑12 18:44:06) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.7']] |
| |  | hl7:entryRelationship
|
| | 0 … * | R | Sub-Observation
The result obtained for an observation may lead to an additional AP observation to refine this result. This sub-observation is again an Anatomic Pathology Observation.
Source: Source: PaLM TF Suppl. APSR 2.0-3
: 6.3.6.7
Contains 1.3.6.1.4.1.19376.1.8.1.4.9 AP Observation Entry (2014‑05‑14 17:09:54) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [@typeCode='COMP'] [hl7:observation [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.9']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  | hl7:entryRelationship
|
| | 0 … * | R | Embedded image.
This observation may carry an illustrative image, either directly embedded or encapsulated within a region of interest.
Source: PaLM TF Suppl. APSR 2.0-3
: 6.3.6.8
Contains 1.3.6.1.4.1.19376.1.8.1.4.10 Embedded Image IHE (2016‑06‑22 16:08:01) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [@typeCode='COMP'] [hl7:observationMedia [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.10']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |  | hl7:entryRelationship
|
| | 0 … * | R | Region of Interest
Contains 2.16.840.1.113883.10.12.307 CDA RegionOfInterest (2005‑09‑07) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [@typeCode='COMP'] [hl7:regionOfInterest [hl7:templateId [@root='2.16.840.1.113883.10.12.307']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP | | | Constraint | This observation may carry an illustrative image, either directly embedded or encapsulated within a region of interest.
In the latter case by an entryRelationship @typeCode="SUBJ" |  | hl7:entryRelationship
|
| | 0 … * | R | Annotation comment
Contains 1.3.6.1.4.1.19376.1.5.3.1.4.2 IHE Comment Entry (2013‑12‑20) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |  | where [hl7:act [hl7:templateId [@root='2.16.840.1.113883.10.20.1.40'] and hl7:templateId [@root='1.3.6.1.4.1.19376.1.5.3.1.4.2']]] |
| |  |  | @typeCode
|
| cs | 0 … 1 | F | COMP | | Included | 1 … * | M | from 1.3.6.1.4.1.19376.1.3.10.9.1 X Specimen Identified (2014‑12‑22 15:54:30)
Specimen identification
| | | Example | Example for specimen referencing <specimen>
<!-- which specimen is used for this observation -->
<specimenRole>
<id root="1.3.6.1.4.1.19376.1.8.1.4.999999" extension="slide_A_1_PR"/> </specimenRole></specimen> |  | hl7:specimen
|
| | 1 … * | M | Required in XD-LAB when more than one specimen is documented at this level. Always required in APSR.
| PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  |  | @typeCode
|
| cs | 0 … 1 | F | SPC |  |  | hl7:specimenRole
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified | | cs | 0 … 1 | F | SPEC | | II | 1 … 1 | R | Specimen ID, coming from the LIS | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  | hl7:reference
|
| | 0 … * | R | Reference to an external document, or to external observations, e.g. concerning molecular pathology or genetic testing report
Contains 2.16.840.1.113883.10.12.324 CDA Reference (2005‑09‑07) | PaLM TF Suppl. APSR 2.0‑3: 6.3.6.7 AP observation |
|
6.3.6.8 Embedded Image – 1.3.6.1.4.1.19376.1.8.1.4.10
6.3.6.8.1 Definition and purpose
This Content Module is usable within an <entry> element, in relationship with a
display anchor carried in the referencedObject attribute of a <renderMultimedia>
element in the <text> element of the <section> holding this <entry>.
The <observationMedia> element carries an image, embedded in B64. This element may be standalone, or encapsulated within a <regionOfInterest> element which defines an overlay
shape to focus on a part of the image.
This <observationMedia> element embeds the image binary data, encoded in B64.
6.3.6.8.2 Specification and Example
6.3.6.9 X Specimen Identified - 1.3.6.1.4.1.19376.1.3.10.9.1
6.3.6.9.1 Definition and purpose
This Specimen Identification <entry> provides the machine-readable links between any type of procedure or observation or embedded image or containers to the specimen, on that this procedure is performed (parent specimen), or from which this observation or image is obtained.
It is required when more than one specimen is documented at this level.
It is only used in PaLM templates for Problem Organizer, Specimen Procedure Steps, Observations, Embedded images, and Containers in Specimen Procedure Steps.
6.3.6.9.2 Specification and Example
6.3.6.10 UICC/AJCC TNM Staging and Grading - 1.3.6.1.4.1.19376.1.3.10.4.2
6.3.6.10.1 Definition and purpose
The UICC/AJCC TNM <entry> Content Modul is a template for a complex structured observation usable in tumor pathology contexts. It contains in machine-readable form all the information concerning a TNM formula for a problem investigated. It is a constraint version of an AP Generic observation <entry> Content Module. It is usable within a Problem Organizer <entry> Content Module only.
This Content Module structures the machine-readable data, which characterize the staging and grading of a malignant tumor according the UICC/AJCC regulations for the TNM system. A series of supporting templates is used for all the detailed descriptions of the elements of the TNM system. Value sets are mentioned for the most recent UICC/AJCC 8th edition and its immediate predecessor 7th edition only.
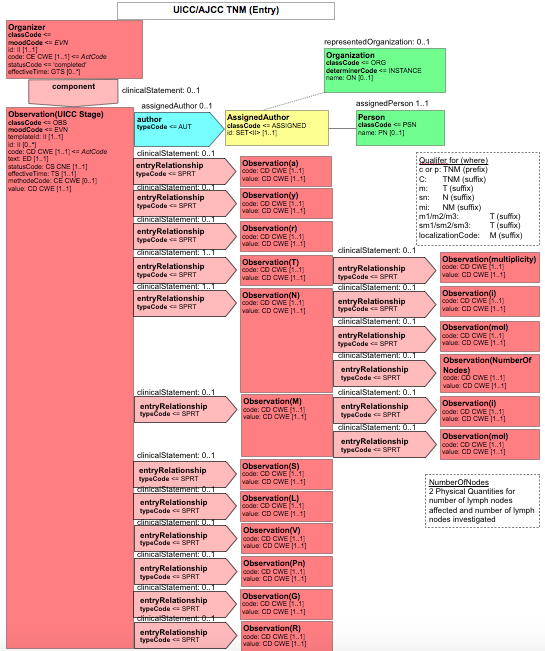
Figure 6.3.6.10.1-1 CDA-R-MIM of the UICC/AJCC TNM <entry>
6.3.6.10.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.3.10.4.2 | Effective Date | valid from 2014‑05‑13 15:45:13 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMStageObservation | Display Name | TNM Stage Observation |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF)
TNM stage with supporting categories |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.4.2 |
|---|
| Label | PaLM Suppl. APSR 2.0‑3: 6.3.6.10 TNM
|
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 1 template, Uses 17 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.1 | Containment |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 1.3.6.1.4.1.19376.1.3.10.9.32 | Containment |  TNM y (2.0) TNM y (2.0) | 2015‑06‑22 17:28:17 | | 1.3.6.1.4.1.19376.1.3.10.9.30 | Containment |  TNM a (2.0) TNM a (2.0) | 2015‑06‑22 15:13:46 | | 1.3.6.1.4.1.19376.1.3.10.9.31 | Containment |  TNM r (2.0) TNM r (2.0) | 2015‑06‑22 17:38:42 | | 1.3.6.1.4.1.19376.1.3.10.9.25 | Containment |  TNM T-Observation (2.0) TNM T-Observation (2.0) | 2014‑12‑02 15:54:17 | | 1.3.6.1.4.1.19376.1.3.10.9.26 | Containment |  TNM N-Observation (2.0) TNM N-Observation (2.0) | 2014‑12‑02 16:34:50 | | 1.3.6.1.4.1.19376.1.3.10.9.27 | Containment |  TNM M-Observation (2.0) TNM M-Observation (2.0) | 2014‑12‑02 16:32:30 | | 1.3.6.1.4.1.19376.1.3.10.9.33 | Containment |  TNM Serum tumor markers (2.0) TNM Serum tumor markers (2.0) | 2015‑11‑06 14:28:44 | | 1.3.6.1.4.1.19376.1.3.10.9.34 | Containment |  TNM Lymphatic Invasion (2.0) TNM Lymphatic Invasion (2.0) | 2015‑11‑16 14:01:58 | | 1.3.6.1.4.1.19376.1.3.10.9.35 | Containment |  TNM Venous Invasion (2.0) TNM Venous Invasion (2.0) | 2015‑11‑16 13:40:28 | | 1.3.6.1.4.1.19376.1.3.10.9.36 | Containment |  TNM Perineural Invasion (2.0) TNM Perineural Invasion (2.0) | 2015‑11‑11 16:13:40 | | 1.3.6.1.4.1.19376.1.3.10.9.28 | Containment |  TNM Grading (2.0) TNM Grading (2.0) | 2015‑11‑15 13:01:18 | | 1.3.6.1.4.1.19376.1.3.10.9.29 | Containment |  TNM R-status (2.0) TNM R-status (2.0) | 2015‑06‑30 13:38:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation classCode="OBS" moodCode="ENV" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.4.2"/> <id root="1.2.3.999" extension="--example only--"/> <code code="21902-2" codeSystem="2.16.840.1.113883.6.1"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20170712152253"/> </effectiveTime> <value xsi:type="CD" code="IIA" displayName="stage IIA" codeSystem="2.16.840.1.113883.15.6"/> <performer>
<!-- template 1.3.6.1.4.1.19376.1.3.3.1.7 'Laboratory Performer' (2008-08-08T00:00:00) -->
</performer> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant> <participant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.7 'Additional participant in an entry APSR (Body)' (2016-07-12T18:44:06) -->
</participant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* M -->
<entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.32 'TNM y' (2015-06-22T17:28:17) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.30 'TNM a ' (2015-06-22T15:13:46) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.31 'TNM r' (2015-06-22T17:38:42) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.25 'TNM T-Observation' (2014-12-02T15:54:17) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.26 'TNM N-Observation' (2014-12-02T16:34:50) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.27 'TNM M-Observation' (2014-12-02T16:32:30) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.33 'TNM Serum tumor markers' (2015-11-06T14:28:44) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.34 'TNM Lymphatic Invasion' (2015-11-16T14:01:58) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.35 'TNM Venous Invasion' (2015-11-16T13:40:28) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.36 'TNM Perineural Invasion' (2015-11-11T16:13:40) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.28 'TNM Grading' (2015-11-15T13:01:18) -->
</entryRelationship> <entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.29 'TNM R-status' (2015-06-30T13:38:30) -->
</entryRelationship></observation> |
|
|
Supportive Template TNM T-Observation - 1.3.6.1.4.1.19376.1.3.10.9.25
Supportive Template TNM N-Observation - 1.3.6.1.4.1.19376.1.3.10.9.26
| Id | 1.3.6.1.4.1.19376.1.3.10.9.26 | Effective Date | valid from 2014‑12‑02 16:34:50 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMNObservation | Display Name | TNM N-Observation |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.26 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 5 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 1.3.6.1.4.1.19376.1.3.10.9.37 | Containment |  TNM Number of nodes (2.0) TNM Number of nodes (2.0) | 2014‑12‑02 16:48:36 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <hl7:observation>
<hl7:templateId/> <hl7:id root="1.2.3.999" extension="--example only--"/> <hl7:code code="21900-6" codeSystem="2.16.840.1.113883.6.1" displayName="Regional lymph nodes.pathology [Class] Cancer"/> <hl7:text/> <hl7:statusCode code="completed"/> <hl7:value xsi:type="CD" code="62455006" codeSystem="2.16.840.1.113883.6.96" displayName="N0 category (finding)">
<hl7:qualifier>
<hl7:name/> <hl7:value code="C1" codeSystem="2.16.840.1.113883.15.6" displayName="C1"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="p" codeSystem="2.16.840.1.113883.15.16" displayName="p"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="i+" codeSystem="2.16.840.1.113883.15.16" displayName="(i+)"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="mol+" codeSystem="2.16.840.1.113883.15.16" displayName="(mol+)"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="sn" codeSystem="2.16.840.1.113883.15.16" displayName="(sn)"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="mi" codeSystem="2.16.840.1.113883.15.16" displayName="(mi)"/> </hl7:qualifier> </hl7:value> <hl7:author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</hl7:author> <hl7:informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</hl7:informant> <hl7:participant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.7 'Additional participant in an entry APSR (Body)' (2016-07-12T18:44:06) -->
</hl7:participant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
<hl7:entryRelationship>
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.37 'TNM Number of nodes' (2014-12-02T16:48:36) -->
</hl7:entryRelationship></hl7:observation> |
|
|
Supportive Template TNM Number of Nodes - 1.3.6.1.4.1.19376.1.3.10.9.37
| Id | 1.3.6.1.4.1.19376.1.3.10.9.37 | Effective Date | valid from 2014‑12‑02 16:48:36 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMNumberOfNodes | Display Name | TNM Number of nodes |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.37 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 3 templates, Uses 4 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.26 | Containment |  TNM N-Observation (2.0) TNM N-Observation (2.0) | 2014‑12‑02 16:34:50 | | 1.3.6.1.4.1.19376.1.3.10.4.2 |  |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | generated example | <hl7:observation>
<hl7:templateId/> <hl7:id root="1.2.3.999" extension="--example only--"/> <hl7:code code="118553007" codeSystem="2.16.840.1.113883.6.96" displayName="Number ratio (attribute)">
<hl7:qualifier>
<hl7:name/> <hl7:value code="441709004" codeSystem="2.16.840.1.113883.6.96" displayName="Specimen from sentinel lymph node (specimen) "/> </hl7:qualifier> </hl7:code> <hl7:text/> <hl7:statusCode code="completed"/> <hl7:effectiveTime>
<hl7:low value="20180122172854"/> </hl7:effectiveTime> <hl7:value xsi:type="RTO_PQ_PQ">
<hl7:numerator value="3" unit="1"/> <hl7:denominator value="10" unit="1"/> </hl7:value> <hl7:author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</hl7:author> <hl7:informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</hl7:informant> <hl7:participant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.7 'Additional participant in an entry APSR (Body)' (2016-07-12T18:44:06) -->
</hl7:participant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
</hl7:observation> |
|
|
Supportive Template TNM M-Observation - 1.3.6.1.4.1.19376.1.3.10.9.27
| Id | 1.3.6.1.4.1.19376.1.3.10.9.27 | Effective Date | valid from 2014‑12‑02 16:32:30 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMMObservation | Display Name | TNM M-Observation |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.27 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 4 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | generated example | <hl7:observation>
<hl7:templateId/> <hl7:id root="1.2.3.999" extension="--example only--"/> <hl7:code code="21901-4" codeSystem="2.16.840.1.113883.6.1"/> <hl7:text/> <hl7:statusCode/> <hl7:effectiveTime>
<hl7:low value="20171129181153"/> </hl7:effectiveTime> <hl7:value xsi:type="CD" code="30893008" codeSystem="2.16.840.1.113883.6.96" displayName="M0 stage (tumor staging)">
<hl7:qualifier>
<hl7:name/> <hl7:value code="84068000" codeSystem="2.16.840.1.113883.6.96" displayName="C1 - TNM certainty factor"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="p" codeSystem="2.16.840.1.113883.15.16" displayName="p"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="i+" codeSystem="2.16.840.1.113883.15.16" displayName="(i+)"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="mol+" codeSystem="2.16.840.1.113883.15.16" displayName="(mol+)"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="PUL" codeSystem="2.16.840.1.113883.15.16" displayName="PUL"/> </hl7:qualifier> <hl7:qualifier>
<hl7:name/> <hl7:value code="mi" codeSystem="2.16.840.1.113883.15.16" displayName="(mi)"/> </hl7:qualifier> </hl7:value> <hl7:author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</hl7:author> <hl7:informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</hl7:informant> <hl7:participant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.7 'Additional participant in an entry APSR (Body)' (2016-07-12T18:44:06) -->
</hl7:participant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 0..1 C -->
</hl7:observation> |
|
|
Supportive Template TNM Grading - 1.3.6.1.4.1.19376.1.3.10.9.28
Supportive Template TNM R-Status - 1.3.6.1.4.1.19376.1.3.10.9.29
| Id | 1.3.6.1.4.1.19376.1.3.10.9.29 | Effective Date | valid from 2015‑06‑30 13:38:30 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMRstatus | Display Name | TNM R-status |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF)
Residual Tumor Status after any therapy |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.29 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 3 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example for R0 | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.29"/> <id root="1.2.3.999" extension="--example only--"/> <code code="445200009 " codeSystem="2.16.840.1.113883.6.96"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20170712150312"/> </effectiveTime> <value xsi:type="CD" code="258254000" displayName="Residual tumor stage R0 (finding)" codeSystem="2.16.840.1.113883.6.96"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* M -->
</observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … 1 | R | | (TNM...tus) |  | @classCode
|
| cs | 1 … 1 | F | OBS |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | @negationInd
|
| bl | 0 … 1 | | |  | hl7:templateId
|
| II | 1 … 1 | M | | (TNM...tus) |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.10.9.29 |  | hl7:id
|
| II | 0 … * | R | | (TNM...tus) |  | hl7:code
|
| CD (extensible) | 1 … 1 | M | | (TNM...tus) |  |  | @code
|
| CONF | 1 … 1 | F | 445200009 |  |  | @codeSystem
|
| 1 … 1 | F | 2.16.840.1.113883.6.96 (SNOMED Clinical Terms) |  |  | @codeSystemName
|
| 1 … 1 | F | SCT |  | hl7:text
|
| ED | 0 … 1 | R | | (TNM...tus) |  | hl7:statusCode
|
| CS (required) | 1 … 1 | M | | (TNM...tus) | | | CONF | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.2 (DYNAMIC) | Error: Cannot find value set "1.3.6.1.4.1.19376.1.3.11.2" |
|
|  | hl7:effectiveTime
|
| IVL_TS | 1 … 1 | M | | (TNM...tus) |  | hl7:value
|
| CD (extensible) | 1 … 1 | R | | (TNM...tus) | | | CONF | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.21 UICC Residual tumor (R) classification (2015‑06‑25 11:23:51) |
|  | hl7:author
|
| | 0 … * | C | Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | (TNM...tus) |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| |  | hl7:informant
|
| | 0 … * | R | Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | (TNM...tus) |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| | | Included | 1 … * | M | from 1.3.6.1.4.1.19376.1.3.10.9.1 X Specimen Identified (2014‑12‑22 15:54:30) |  | hl7:specimen
|
| | 1 … * | M | Required in XD-LAB when more than one specimen is documented at this level. Always required in APSR.
| PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  |  | @typeCode
|
| cs | 0 … 1 | F | SPC |  |  | hl7:specimenRole
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified | | cs | 0 … 1 | F | SPEC | | II | 1 … 1 | R | Specimen ID, coming from the LIS | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |
|
Supportive Template TNM a - 1.3.6.1.4.1.19376.1.3.10.9.30
| Id | 1.3.6.1.4.1.19376.1.3.10.9.30 | Effective Date | valid from 2015‑06‑22 15:13:46 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMa | Display Name | TNM a |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.30 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation negationInd="false" classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.30"/> <code code="277646006" codeSystem="2.16.840.1.113883.6.96" displayName="At autopsy (tumor staging)"/> <text/> <statusCode code="completed"/> <value xsi:type="CD" code="a" displayName="a" codeSystem="2.16.840.1.113883.15.6"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant></observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … 1 | R | | (TNMa) |  | @classCode
|
| cs | 1 … 1 | F | OBS |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | @negationInd
|
| bl | 0 … 1 | | |  | hl7:templateId
|
| II | 1 … 1 | M | | (TNMa) |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.10.9.30 |  | hl7:id
|
| II | 0 … * | R | | (TNMa) |  | hl7:code
|
| CE (extensible) | 1 … 1 | M | additional descriptors:
a - (tumor staging) at autopsy | (TNMa) | | | CONF | | @code shall be "277646006" | | @codeSystem shall be "2.16.840.1.113883.6.96" | | @codeSystemName shall be "SNOMED CT" | | @displayName shall be "At autopsy (tumor staging)" | | or | | @code shall be "85699-7" | | @codeSystem shall be "2.16.840.1.113883.6.1" | | @codeSystemName shall be "LOINC" | | @displayName shall be "Autopsy was performed" |
|  | hl7:text
|
| ED | 0 … 1 | R | | (TNMa) |  | hl7:statusCode
|
| CS (required) | 1 … 1 | M | | (TNMa) | | | CONF | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.2 (DYNAMIC) | Error: Cannot find value set "1.3.6.1.4.1.19376.1.3.11.2" |
|
|  | hl7:effectiveTime
|
| IVL_TS | 1 … 1 | M | | (TNMa) |  | hl7:value
|
| CD (extensible) | 1 … 1 | R | | (TNMa) | | | CONF | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.41 UICC Autopsy (2015‑11‑18 15:55:17) |
|  | hl7:author
|
| II | 0 … * | C | Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | (TNMa) |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| |  | hl7:informant
|
| II | 0 … * | R | Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | (TNMa) |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| |
|
Supportive Template TNM r - 1.3.6.1.4.1.19376.1.3.10.9.31
| Id | 1.3.6.1.4.1.19376.1.3.10.9.31 | Effective Date | valid from 2015‑06‑22 17:38:42 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMr | Display Name | TNM r |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.31 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.31"/> <id root="1.2.3.999" extension="--example only--"/> <code code="277647002" codeSystem="2.16.840.1.113883.6.96" displayName="Tumor recurrence (tumor staging)"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20170712103938"/> </effectiveTime> <value xsi:type="CD" code="r" displayName="r" codeSystem="2.16.840.1.113883.15.6"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-06-21T14:02:11) -->
</informant></observation> |
|
|
Supportive Template TNM y - 1.3.6.1.4.1.19376.1.3.10.9.32
| Id | 1.3.6.1.4.1.19376.1.3.10.9.32 | Effective Date | valid from 2015‑06‑22 17:28:17 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMy | Display Name | TNM y |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.32 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation negationInd="false" classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.32"/> <id root="1.2.3.999" extension="--example only--"/> <code code="277649004" codeSystem="2.16.840.1.113883.6.96" displayName="y - During therapy/following treatment (tumor staging)"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20170712105323"/> </effectiveTime> <value xsi:type="CD" code="y" displayName="y" codeSystem="2.16.840.1.113883.15.6"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant></observation> |
|
|
Supportive Template TNM Serum Tumor Markers - 1.3.6.1.4.1.19376.1.3.10.9.33
| Id | 1.3.6.1.4.1.19376.1.3.10.9.33 | Effective Date | valid from 2015‑11‑06 14:28:44 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMserumtumormarker | Display Name | TNM Serum tumor markers |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.33 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 3 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 2.16.840.1.113883.10.12.300 | Containment |  CDA Clinical Statement CDA Clinical Statement | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.33"/> <Id root="1.2.3.999" extension="--example only--"/> <code code="tnmS" codeSystem="1.2.276.0.76.3.1.131.1.5.1"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20170712150818"/> </effectiveTime> <value xsi:type="CD" code="S0" displayName="S0" codeSystem="2.16.840.1.113883.15.6"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant> <entryRelationship typeCode="COMP">
<!-- template 2.16.840.1.113883.10.12.300 'CDA Clinical Statement' (2005-09-07T00:00:00) -->
</entryRelationship></observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … 1 | C | | (TNM...ker) |  | @classCode
|
| cs | 1 … 1 | F | OBS |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | @negationInd
|
| bl | 0 … 1 | | | | | Constraint | In case of male testicular tumor only. Information comes from clinical information or from a clinical statement or from ICD-O-3-Topography (C62.0, C62.1, C62.9) and ICD-O-3 Typing (M9061-M9085). |  | hl7:templateId
|
| II | 1 … 1 | M | | (TNM...ker) |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.10.9.33 |  | hl7:Id
|
| II | 0 … * | R | | (TNM...ker) |  | hl7:code
|
| CE (extensible) | 1 … 1 | M | | (TNM...ker) |  |  | @codeSystem
|
| oid | 1 … 1 | F | 1.2.276.0.76.3.1.131.1.5.1 |  |  | @code
|
| cs | 1 … 1 | F | tnmS |  | hl7:text
|
| ED | 0 … 1 | R | | (TNM...ker) |  | hl7:statusCode
|
| CS (required) | 1 … 1 | M | | (TNM...ker) | | | CONF | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.2 (DYNAMIC) | Error: Cannot find value set "1.3.6.1.4.1.19376.1.3.11.2" |
|
|  | hl7:effectiveTime
|
| IVL_TS | 1 … 1 | M | | (TNM...ker) |  | hl7:value
|
| CD (extensible) | 1 … 1 | R | | (TNM...ker) | | | CONF | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.32 UICC Serum Tumor Markers (2015‑11‑08 18:26:39) |
|  | hl7:author
|
| | 0 … * | C | Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | (TNM...ker) |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| |  | hl7:informant
|
| | 0 … * | R | Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | (TNM...ker) |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| |  | hl7:entryRelationship
|
| | 0 … * | C | Information about clinical laboratory values
Contains 2.16.840.1.113883.10.12.300 CDA Clinical Statement (2005‑09‑07) | (TNM...ker) |  | where [@typeCode='COMP'] [hl7:act [hl7:templateId [@root='2.16.840.1.113883.10.12.301']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.302']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.303']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.304']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.305']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.306']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.307']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.308']] or
[hl7:templateId [@root='2.16.840.1.113883.10.12.309']]] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | COMP |
|
Supportive Template TNM Lymphatic Invasion - 1.3.6.1.4.1.19376.1.3.10.9.34
| Id | 1.3.6.1.4.1.19376.1.3.10.9.34 | Effective Date | valid from 2015‑11‑16 14:01:58 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMLymphaticInvasion | Display Name | TNM Lymphatic Invasion |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.34 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 3 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.34"/> <id root="1.2.3.999" extension="--example only--"/> <code code="395715009" codeSystem="2.16.840.1.113883.6.96" displayName="Status of lymphatic (small vessel) invasion by tumor (observable entity)"/> <statusCode code="completed"/> <effectiveTime>
<low value="20170712144109"/> </effectiveTime> <value xsi:type="CD" code="L0" displayName="no lymphatic invasion" codeSystem="2.16.840.1.113883.15.6"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
</observation> |
|
|
Supportive Template TNM Venous Invasion - 1.3.6.1.4.1.19376.1.3.10.9.35
| Id | 1.3.6.1.4.1.19376.1.3.10.9.35 | Effective Date | valid from 2015‑11‑16 13:40:28 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TNMVenousInvasion | Display Name | TNM Venous Invasion |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.35 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 3 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <hl7:observation>
<hl7:templateId/> <hl7:id root="1.2.3.999" extension="--example only--"/> <hl7:code code="371493002" codeSystem="2.16.840.1.113883.6.96" displayName="Status of venous (large vessel) invasion by tumor (observable entity)"/> <hl7:text/> <hl7:statusCode/> <hl7:effectiveTime>
<hl7:low value="20180122181318"/> </hl7:effectiveTime> <hl7:value xsi:type="CD" code="V0" codeSystem="2.16.840.1.113883.15.16" displayName="V0"/> <hl7:author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</hl7:author> <hl7:informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</hl7:informant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
</hl7:observation> |
|
|
Supportive Template TNM Perineurial Invasion - 1.3.6.1.4.1.19376.1.3.10.9.36
| Id | 1.3.6.1.4.1.19376.1.3.10.9.36 | Effective Date | valid from 2015‑11‑11 16:13:40 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | TnmPerineuralInvasion | Display Name | TNM Perineural Invasion |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.36 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 3 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | Generated example | <observation classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.36"/> <id root="1.2.3.999" extension="--example only--"/> <code code="tnmPn" codeSystem="1.2.276.0.76.3.1.131.1.5.1"/> <originalText/> <statusCode code="completed"/> <value code="Pn0" displayName="Pn0" codeSystem="2.16.840.1.113883.15.6"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <informant>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.6 'CDA Informant(Header&Body) APSR2' (2016-07-08T11:22:58) -->
</informant> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
</observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 0 … 1 | R | | (Tnm...ion) |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | @classCode
|
| cs | 1 … 1 | F | OBS |  | hl7:templateId
|
| II | 1 … 1 | M | | (Tnm...ion) |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.10.9.36 |  | hl7:id
|
| II | 0 … * | R | | (Tnm...ion) |  | hl7:code
|
| CD (extensible) | 1 … 1 | M | | (Tnm...ion) |  |  | @codeSystem
|
| oid | 1 … 1 | F | 1.2.276.0.76.3.1.131.1.5.1 |  |  | @code
|
| cs | 1 … 1 | F | tnmPn |  | hl7:originalText
|
| ED | 0 … 1 | R | | (Tnm...ion) |  | hl7:statusCode
|
| CS (required) | 1 … 1 | M | | (Tnm...ion) | | | CONF | The value of @code shall be drawn from value set 1.3.6.1.4.1.19376.1.3.11.2 (DYNAMIC) | Error: Cannot find value set "1.3.6.1.4.1.19376.1.3.11.2" |
|
|  | hl7:value
|
| CD (extensible) | 1 … 1 | R | | (Tnm...ion) | | | CONF | | The value of @code should be drawn from value set 1.3.6.1.4.1.19376.1.3.11.25 UICC Perineural Invasion (2015‑11‑04 16:20:42) |
|  | hl7:author
|
| II | 0 … * | C | Contains 1.3.6.1.4.1.19376.1.8.1.4.2 CDA author IHE (2016‑06‑21 14:02:11) | (Tnm...ion) |  | where [hl7:author [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.2']]] |
| |  | hl7:informant
|
| II | 0 … * | R | Contains 1.3.6.1.4.1.19376.1.8.1.4.6 CDA Informant(Header&Body) APSR2 (2016‑07‑08 11:22:58) | (Tnm...ion) |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.8.1.4.6']] |
| | | Included | 1 … * | R | from 1.3.6.1.4.1.19376.1.3.10.9.1 X Specimen Identified (2014‑12‑22 15:54:30) |  | hl7:specimen
|
| | 1 … * | R | Required in XD-LAB when more than one specimen is documented at this level. Always required in APSR.
| PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |  |  | @typeCode
|
| cs | 0 … 1 | F | SPC |  |  | hl7:specimenRole
|
| | 1 … 1 | M | | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified | | cs | 0 … 1 | F | SPEC | | II | 1 … 1 | R | Specimen ID, coming from the LIS | PaLM Suppl. APSR 2.0‑3: 6.3.6.9 X Specimen identified |
|
6.3.6.11 ICD-O-3 Typing and Grading - 1.3.6.1.4.1.19376.1.3.10.4.3
6.3.6.11.1 Definition and purpose
The ICD-O-3 Typing and Grading<entry> Content Modul is a template for a complex observation usable in tumor pathology contexts. It contains in machine-readable form all the information concerning ICD-O-3 Typing and Grading for a problem investigated. It is a constraint version of an AP Generic observation <entry> Content Module. It is usable within a Problem Organizer <entry> Content Module only.
This Content Module structures the machine-readable data, which characterize the typing and grading of a tumor or a systemic malignancy according the WHO regulations for the ICD-O-3 system. The supporting template ICD-O-Differentiation is used for a more detailed descriptions of those elements of the ICD-O-3 system. The supporting template ICD-O-Behavior is only to use if the pathologists´ behavior classification is (rarely) deviant from the 5th digit in morphology and behavior code list, and is therefore overriding that digit.
Morphology and topography axes of ICD-O-3 are not combined to a singular, common code, but used simultaneously. A complete ICD-O-3 coding consists always of topography and morphology and behavior codes, displayed together in one line in the human readable section. Therefore, the topography axis of ICD-O-3 SHOULD be coded in a supportive template ICD-O-Topography, specialized from a generic observation, within the same Problem Organizer <entry> Content Module.
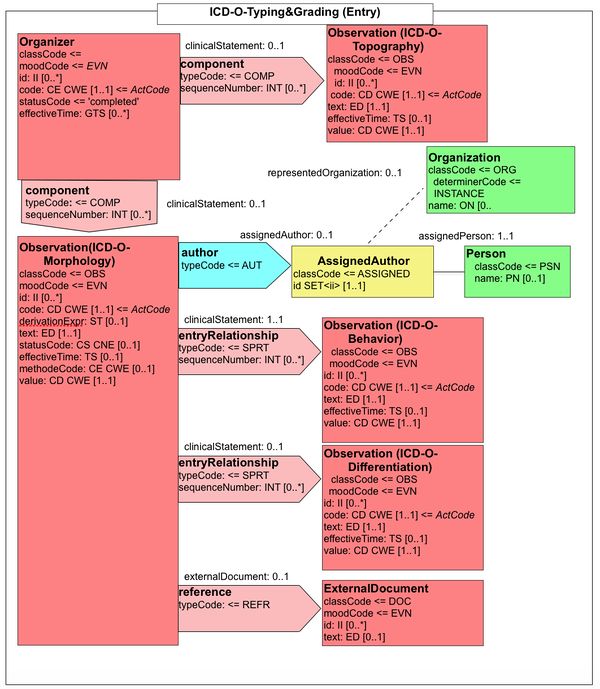
Figure 6.3.6.11.1-1 CDA-R-MIM of the ICD-O-3 <entry>
6.3.6.11.2 Specification and Example
| Id | 1.3.6.1.4.1.19376.1.3.10.4.3 | Effective Date | valid from 2015‑11‑29 17:03:18 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ICD-O-3TypingGrading | Display Name | ICD-O-3 Typing and Grading Observation |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF)
Entry template for observation on ICD-O-3 Typing and Grading |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.4.3 |
|---|
| Label | PaLM Suppl. APSR 2.0‑3: 6.3.6.11 ICD‑O‑3
|
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 0 templates, Uses 9 templates | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.4.2 | Containment |  CDA author IHE CDA author IHE | 2016‑06‑21 14:02:11 | | 1.3.6.1.4.1.19376.1.8.1.4.6 | Containment |  CDA Informant(Header&Body) APSR2 (2.0) CDA Informant(Header&Body) APSR2 (2.0) | 2016‑07‑08 11:22:58 | | 1.3.6.1.4.1.19376.1.8.1.4.7 | Containment |  Additional participant in an entry APSR (Body) Additional participant in an entry APSR (Body) | 2016‑07‑12 18:44:06 | | 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 1.3.6.1.4.1.19376.1.3.10.9.38 | Containment |  ICD-O-3 Behavior (2.0) ICD-O-3 Behavior (2.0) | 2015‑12‑03 09:11:54 | | 1.3.6.1.4.1.19376.1.3.10.9.39 | Containment |  ICD-O-3 Differentiation (2.0) ICD-O-3 Differentiation (2.0) | 2015‑12‑09 17:12:30 | | 1.3.6.1.4.1.19376.1.3.10.4.4 | Containment |  Assessment Scales Observation for Scoring Systems APSR2 (2.0) Assessment Scales Observation for Scoring Systems APSR2 (2.0) | 2016‑10‑06 13:12:00 | | 2.16.840.1.113883.10.12.324 | Containment |  CDA Reference CDA Reference | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | example for use case #1 | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.4.3"/> <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_ICDO"/> <code code="31205-8" codeSystem="2.16.840.1.113883.6.1" displayName="Histology ICD-O-3 Cancer"/> <statusCode code="completed"/> <effectiveTime>
<low value="201001041405-0500"/> </effectiveTime> <value xsi:type="CD" code="8500/3" codeSystem="2.16.840.1.113883.6.43.1"/> <author>
<!-- template 1.3.6.1.4.1.19376.1.8.1.4.2 'CDA author IHE' (2016-06-21T14:02:11) -->
</author> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
<specimen typeCode="SPC">
<specimenRole classCode="SPEC">
<id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_slide_A1_HE"/> </specimenRole> </specimen> <entryRelationship typeCode="SPRT">
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.38 'ICD-O-3 Behavior' (2015-12-03T09:11:54), not used in use case #1 -->
</entryRelationship> <entryRelationship typeCode="SPRT">
<!-- template 1.3.6.1.4.1.19376.1.3.10.9.39 'ICD-O-3 Differentiation' (2015-12-09T17:12:30) -->
</entryRelationship> <entryRelationship typeCode="COMP">
<!-- template 1.3.6.1.4.1.19376.1.3.10.4.4 'Assessment Scales Observation for Scoring Systems APSR2' (2016-10-06T13:12:00) -->
</entryRelationship> <reference>
<!-- template 2.16.840.1.113883.10.12.324 'CDA Reference' (2005-09-07T00:00:00) -->
</reference></observation> |
|
|
Supportive Template ICD-O-Behavior - 1.3.6.1.4.1.19376.1.3.10.9.38
| Id | 1.3.6.1.4.1.19376.1.3.10.9.38 | Effective Date | valid from 2015‑12‑03 09:11:54 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ICD-O-3Behavior | Display Name | ICD-O-3 Behavior |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF)
Behavior code |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.38 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 1 template, Uses 1 template | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.3 | Containment |  ICD-O-3 Typing and Grading Observation (2.0) ICD-O-3 Typing and Grading Observation (2.0) | 2015‑11‑29 17:03:18 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | generated example | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.38"/> <id root="1.2.3.999" extension="--example only--"/> <code code="31206-6" codeSystem="2.16.840.1.113883.6.1" displayName="Behavior ICD-O-3 Cancer"/> <statusCode code="completed"/> <effectiveTime>
<low value="20170305122607"/> </effectiveTime> <value xsi:type="CV" code="3" displayName="/3" codeSystem="2.16.840.1.113883.6.43.1"/> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
</observation> |
|
|
Supportive Template ICD-O-Differentiation - 1.3.6.1.4.1.19376.1.3.10.9.39
| Id | 1.3.6.1.4.1.19376.1.3.10.9.39 | Effective Date | valid from 2015‑12‑09 17:12:30 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ICD-O-3Differentiation | Display Name | ICD-O-3 Differentiation |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF)
Differentiation, sive grading code |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.39 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 1 template, Uses 2 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.3 | Containment |  ICD-O-3 Typing and Grading Observation (2.0) ICD-O-3 Typing and Grading Observation (2.0) | 2015‑11‑29 17:03:18 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 2.16.840.1.113883.10.12.300 | Containment |  CDA Clinical Statement CDA Clinical Statement | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | for use case #1 | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.39"/> <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_ICDO_D"/> <code code="21858-6" codeSystem="2.16.840.1.113883.6.1" displayName="Grade Cancer"/> <statusCode code="completed"/> <value xsi:type="CD" code="1" displayName="1" codeSystem="2.16.840.1.113883.6.43.1"/> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
<specimen typeCode="SPC">
<specimenRole classCode="SPEC">
<id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_slide_A1_HE"/> </specimenRole> </specimen> <entryRelationship typeCode="EVN">
<!-- template 2.16.840.1.113883.10.12.300 'CDA Clinical Statement' (2005-09-07T00:00:00) -->
</entryRelationship></observation> |
|
|
Supportive Template ICD-O-Topography - 1.3.6.1.4.1.19376.1.3.10.9.41
| Id | 1.3.6.1.4.1.19376.1.3.10.9.41 | Effective Date | valid from 2015‑11‑16 18:38:07 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ICD-O-3Topography | Display Name | ICD-O-3 Topography |
|---|
| Description | Template CDA Observation (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.41 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 0 templates, Uses 1 template | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | example for use case #1 | <observation classCode="OBS" moodCode="EVN" negationInd="false">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.41"/> <id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_ICDO_T"/> <code code="42129-7" codeSystem="2.16.840.1.113883.6.1" displayName="Site coding system.current"/> <statusCode code="completed"/> <effectiveTime>
<low value="201001041405-0500"/> </effectiveTime> <value xsi:type="CV" code="C50.4" displayName="Breast, upper outer quadrant" codeSystem="2.16.840.1.113883.6.43.1">
<qualifier>
<name code="20228-3" codeSystem="2.16.840.1.113883.6.1" displayName="Anatomic part Laterality"/> <value value="L" codeSystem="2.16.840.1.113883.6.1" displayName="Left"/> </qualifier> </value> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
<specimen typeCode="SPC">
<specimenRole classCode="SPEC">
<id root="1.3.6.1.4.1.19376.1.8.9.1" extension="A7102400008_slide_A1_HE"/> </specimenRole> </specimen></observation> |
|
|
6.3.6.12 Assessment Scales for Scoring systems - 1.3.6.1.4.1.19376.1.3.10.4.4
6.3.6.12.1 Definition and purpose
The Assessment Scales <entry> Content Modul is a template for a complex observation, especially usable in tumor pathology contexts, within a Problem organizer or as sub-observation for grading observations. It contains in machine-readable form all the information concerning (semi)quantitative scoring systems often used for describing a tumor grading for a problem investigated. It is a constraint version of an AP Generic observation <entry> Content Module. It is usable within the ICD-O-3 Typing and Grading <entry> or within a Problem Organizer <entry> Content Module.
This Content Module structures the machine-readable data, which characterize the grading components of a tumor or a systemic disease according specific regulations.
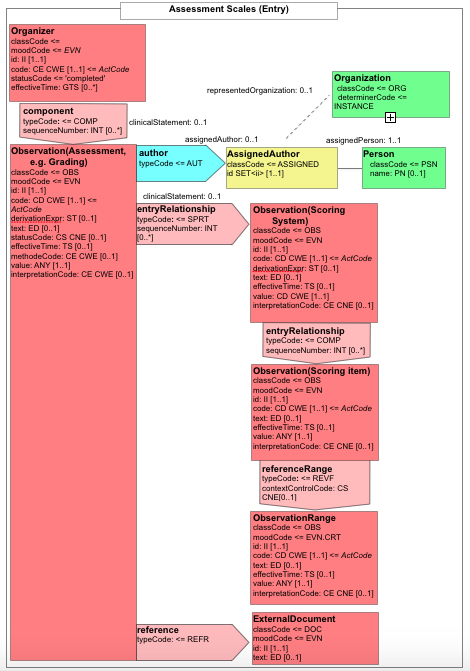
Figure 6.3.6.12.1-1 CDA-R-MIM of an Assessment Scales <entry>
6.3.6.12.2 Specification and Example
Assessment Scales Observation for Scoring Systems APSR2 - 1.3.6.1.4.1.19376.1.3.10.4.4
Supportive Template Assessment Scoring System - 1.3.6.1.4.1.19376.1.3.10.9.42
| Id | 1.3.6.1.4.1.19376.1.3.10.9.42 | Effective Date | valid from 2016‑10‑10 11:33:36 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | AssessmentScoringSystemAPSR2.0 | Display Name | Assessment Scoring System APSR 2.0 |
|---|
| Description | Supportive Observation entry for describing a scoring system |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.10.9.42 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 2 templates, Uses 4 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.4.4 | Containment |  Assessment Scales Observation for Scoring Systems APSR2 (2.0) Assessment Scales Observation for Scoring Systems APSR2 (2.0) | 2016‑10‑06 13:12:00 | | 1.3.6.1.4.1.19376.1.3.10.4.3 |  |  ICD-O-3 Typing and Grading Observation (2.0) ICD-O-3 Typing and Grading Observation (2.0) | 2015‑11‑29 17:03:18 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.10.9.1 | Include |  X Specimen Identified X Specimen Identified | 2014‑12‑22 15:54:30 | | 1.3.6.1.4.1.19376.1.3.10.9.43 | Containment |  Assessment Scoring Item APSR 2.0 (2.0) Assessment Scoring Item APSR 2.0 (2.0) | 2016‑10‑11 10:26:17 | | 2.16.840.1.113883.10.12.324 | Containment |  CDA Reference CDA Reference | 2005‑09‑07 | | 2.16.840.1.113883.10.12.329 | Containment |  CDA Precondition CDA Precondition | 2005‑09‑07 |
|
|
|---|
| Relationship | Specialization: template 1.3.6.1.4.1.19376.1.8.1.4.9 (2014‑05‑14 17:09:54) |
|---|
| Example | | generated example for use case #1, ER-Allred Score | <observation negationInd="false" classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.10.9.42"/> <id root="1.2.3.999" extension="--example only--"/> <code code="278061009 | Scores (qualifier value) |" codeSystem="2.16.840.1.113883.6.96" displayName="Score"/> <derivationExpr text="sum"/> <text/> <statusCode code="completed"/> <effectiveTime>
<low value="20170224134516"/> </effectiveTime> <value code="445104009" codeSystem="2.16.840.1.113883.6.96" displayName="Sum of proportion of positive staining neoplastic cells score and average intensity of staining score for hormone receptors using immunohistochemistry (observable entity)"/> <methodCode code="0107" displayName="Microscopy" codeSystem="2.16.840.1.113883.5.84"/> <!-- include template 1.3.6.1.4.1.19376.1.3.10.9.1 'X Specimen Identified' (2014-12-22T15:54:30) 1..* R -->
<entryRelationship>
<sequenceNumber value="1"/> <seperatableInd value="false"/> <!-- template 1.3.6.1.4.1.19376.1.3.10.9.43 'Assessment Scoring Item APSR 2.0' (2016-10-11T10:26:17) -->
</entryRelationship> <reference>
<!-- template 2.16.840.1.113883.10.12.324 'CDA Reference' (2005-09-07T00:00:00) -->
</reference> <precondition>
<!-- template 2.16.840.1.113883.10.12.329 'CDA Precondition' (2005-09-07T00:00:00) -->
</precondition> <referenceRange typeCode="REFV">
<observationRange classCode="OBS" moodCode="EVN.CRT">
<code code="--code--" codeSystem="2.16.840.1.113883.5.4"/> <text/> <value xsi:type="ANY"/> <interpretationCode code="EXP" displayName="Expected" codeSystem="2.16.840.1.113883.5.83"/> </observationRange> </referenceRange></observation> |
|
|
Supportive Template Assessment Scoring Item - 1.3.6.1.4.1.19376.1.3.10.9.43
6.3.6.13 Participant Pertinent Insurance Information - 1.3.6.1.4.1.19376.1.3.10.2.5
6.3.6.13.1 Definition and purpose
This participant is carrying key information of the insurance status of the patient, as it may be requested e.g. by cancer registries. It is used in a header only. It is not intended to provide the whole information about payment details.
6.3.6.13.2 Specification and Example
6.3.7 External Content Modules referenced by APSR 2.0
6.3.7.1 Definition and purpose
For some of the templates described above supportive templates are used, defined and specified by XD-LAB or PCC. They are listed in table 5.5-1 (see above), and shown here for better understanding only.
6.3.7.2 Specifications and Examples
IHE Reason for Referral Sub-Section - 1.3.6.1.4.1.19376.1.5.3.1.3.1
History of Present Illness Sub-Section - 1.3.6.1.4.1.19376.1.5.3.1.3.4
CDA Active Problems Sub-Section - 1.3.6.1.4.1.19376.1.5.3.1.3.6
Specimen Received Child element - 1.3.6.1.4.1.19376.1.3.1.3
Laboratory Observation Child element - 1.3.6.1.4.1.19376.1.3.1.6
| Id | 1.3.6.1.4.1.19376.1.3.1.6 ref (from repository: XDLAB-) | Effective Date | valid from 2008‑08‑08 |
|---|
| Status |  Active Active | Version Label | 2017 |
|---|
| Name | LaboratoryObservation | Display Name | Laboratory Observation |
|---|
| Description | The document SHALL contain at least one Laboratory Observation (See REFERENCE) under the Specimen Act of each Laboratory Data Processing Entry (See REFERENCE). The Laboratory Observation SHALL record a single laboratory observation in the document, either standalone or as part of a battery. It should be distinguished from the PCC Simple Observation
template. |
|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.3.1.6 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 8 templates, Uses 6 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.3.6 | Containment |  Problem Organizer (2.0) Problem Organizer (2.0) | 2015‑08‑13 10:24:55 | | 1.3.6.1.4.1.19376.1.3.10.3.1 |  |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | 1.3.6.1.4.1.19376.1.8.1.2.1 |  |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 |  |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 |  |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 |  |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.8.1.2.5 |  |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | Uses | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.3.1.2.1 | Containment |  Non-Human Subject (2017) Non-Human Subject (2017) | DYNAMIC | | 1.3.6.1.4.1.19376.1.3.3.1.3.1 | Containment |  Human Patient with Non-Human Subject (2017) Human Patient with Non-Human Subject (2017) | DYNAMIC | | 1.3.6.1.4.1.19376.1.3.3.1.7 | Containment |  Laboratory Performer (2017) Laboratory Performer (2017) | DYNAMIC | | 1.3.6.1.4.1.19376.1.3.10.9.17 | Containment |  Author with Name, Addr, Telecom (2017) Author with Name, Addr, Telecom (2017) | DYNAMIC | | 1.3.6.1.4.1.19376.1.5.3.1.4.2 | Containment |  IHE Comment Entry (2014) IHE Comment Entry (2014) | DYNAMIC | | 1.3.6.1.4.1.19376.1.3.1.2 | Containment |  Specimen Collection (2017) Specimen Collection (2017) | DYNAMIC |
|
|
|---|
| Relationship | Generalization: template 2.16.840.1.113883.10.12.303 (2005‑09‑07) |
|---|
| Example | | Example | <observation classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.1.6"/> <id root=" " extension=" "/> <code code="10912-4" codeSystem="2.16.840.1.113883.6.1" codeSystemName="LOINC" displayName="Lead Measurement, Quantitative">
<originalText>
<reference value="pbTest"/> </originalText> </code> <statusCode code="completed"/> <effectiveTime value="20071108000000.0000-0500"/> <value xsi:type="PQ" unit="mg/dL" value="6.4"/> <interpretationCode code="H" codeSystem="2.16.840.1.113883.5.83"/> <methodCode code=" " codeSystem=" " codeSystemName=" "/> <specimen typeCode="SPC">
<specimenRole classCode="SPEC">
<id extension="555847399003" root="1.3.6.1.4.1.19376.1.3.4"/> <specimenPlayingEntity>
<code code="BLD" codeSystem="2.16.840.1.113883.5.129" codeSystemName="HL7" displayName="Blood"/> </specimenPlayingEntity> </specimenRole> </specimen> <performer typeCode="PRF">
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.7"/> <!-- ... -->
</performer> <author typeCode="AUT">
<!-- ... -->
</author> <participant typeCode="AUTHEN">
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.5"/> <!-- ... -->
</participant> <participant typeCode="RESP">
<!-- ... -->
</participant> <participant typeCode="DEV">
<!-- ... -->
</participant> <entryRelationship typeCode="COMP">
<act classCode="ACT" moodCode="EVN">
<templateId root="2.16.840.1.113883.10.20.1.40"/> <templateId root="1.3.6.1.4.1.19376.1.5.3.1.4.2"/> <code code="48767-8" codeSystem="2.16.840.1.113883.6.1" codeSystemName="LOINC" displayName="Annotation Comment"/> <text>
<reference value="pbTestComment"/> </text> <statusCode code="completed"/> </act> </entryRelationship> <entryRelationship typeCode="REFR">
<sequenceNumber value="1"/> <observation classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.1.6"/> <code code="10912-4" codeSystem="2.16.840.1.113883.6.1" codeSystemName="LOINC" displayName="Lead Measurement, Quantitative">
<originalText>
<reference value="pbTest"/> </originalText> </code> <statusCode code="completed"/> <effectiveTime value="20071101000000.0000-0500"/> <value xsi:type="PQ" unit="mg/dL" value="8.7"/> <interpretationCode code="H" codeSystem="2.16.840.1.113883.5.83"/> <entryRelationship typeCode="COMP">
<act classCode="ACT" moodCode="EVN">
<templateId root="2.16.840.1.113883.10.20.1.40"/> <templateId root="1.3.6.1.4.1.19376.1.5.3.1.4.2"/> <code code="48767-8" codeSystem="2.16.840.1.113883.6.1" codeSystemName="LOINC" displayName="Annotation Comment"/> <text>
<reference value="pbTestComment"/> </text> <statusCode code="completed"/> </act> </entryRelationship> </observation> </entryRelationship> <referenceRange typeCode="REFV">
<observationRange classCode="OBS" moodCode="EVN.CRT">
<value xsi:type="IVL_PQ">
<low value="0.0" unit="mg/dL"/> <high value="5.0" unit="mg/dL"/> </value> <interpretationCode code="N" codeSystem="2.16.840.1.113883.5.83"/> </observationRange> <lab:precondition typeCode="PRCN">
<lab:criterion classCode="COND">
<lab:code code="AGE"/> <lab:value xsi:type="IVL_PQ">
<lab:low value="35" unit="Y"/> <lab:high value="55" unit="Y"/> </lab:value> </lab:criterion> </lab:precondition> </referenceRange></observation> |
|
| Example | | Example | <observation classCode="OBS" moodCode="EVN">
<templateId root="1.3.6.1.4.1.19376.1.3.1.3"/> <code code="11273-0" codeSystem="2.16.840.1.113883.6.1" codeSystemName="LOINC" displayName="ERYTHROCYTES"/> <statusCode code="completed"/> <effectiveTime value="20060321063000.0000-0500"/> <value xsi:type="PQ" value="4.95" unit="10*6/mm3"/> <interpretationCode code="N" codeSystem="2.16.840.1.113883.5.83"/> <entryRelationship typeCode="REFR">
<!-- Previous result 4.85 from Mar 12, 2006 08:15 -->
<observation classCode="OBS" moodCode="EVN">
<code code="11273-0" codeSystem="2.16.840.1.113883.6.1"/> <statusCode code="completed"/> <effectiveTime value="20060312081500.0000-0500"/> <value xsi:type="PQ" value="4.85" unit="10*6/mm3"/> </observation> </entryRelationship> <!-- reference range given patient sex -->
<referenceRange typeCode="REFV">
<observationRange classCode="OBS" moodCode="EVN.CRT">
<value xsi:type="IVL_PQ">
<low value="4.50" unit="10*6/mm3"/> <high value="6.00" unit="10*6/mm3"/> </value> </observationRange> <lab:precondition typeCode="PRCN">
<lab:criterion classCode="COND">
<lab:code code="SEX"/> <lab:value xsi:type="CD" code="M" codeSystem="2.16.840.1.113883.5.1"/> </lab:criterion> </lab:precondition> </referenceRange></observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 1 … 1 | M | | (Lab...ion) |  | @classCode
|
| cs | 1 … 1 | F | OBS |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | hl7:templateId
|
| II | 1 … 1 | M | | (Lab...ion) |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.1.6 |  | hl7:id
|
| II | 0 … 1 | | A Laboratory Observation identifier MAY be present. | (Lab...ion) |  | hl7:code
|
| CD | 1 … 1 | R | Unique test code in an international standard (LOINC or SNOMED CT) or a national standard (e.g. JC10 in Japan) | (Lab...ion) |  | hl7:statusCode
|
| CS | 1 … 1 | M | ‘completed’ when the result is present.
‘aborted’ if the test could not be performed. | (Lab...ion) | | | CONF | | @code shall be "completed" | | or | | @code shall be "aborted" |
|  | hl7:effectiveTime
|
| IVL_TS | 0 … 1 | R | Physiologically relevant time | (Lab...ion) |  | hl7:value
|
| ANY | 0 … 1 | R | The result obtained for this test using the appropriate data type. Numeric results use data type PQ, which includes the unit. The result is absent in case of ‘obsolete’ or ‘aborted’ observation. | (Lab...ion) |  | hl7:interpretationCode
|
| CE | 0 … 1 | R | One or more codes interpreting the result, expressed with ObservationInterpretation vocabulary (e.g. H = high, L = low)
In case of a antimicrobial susceptibility test in microbiology, the vocabulary domain is ObservationInterpretationSusceptibility: S = susceptible, R = resistant, I = intermediate, VS = very susceptible, MS = moderately susceptible | (Lab...ion) |  | hl7:methodCode
|
| CE | 0 … 1 | R | method used for this observation expressed with ObservationMethod vocabulary (CWE) | (Lab...ion) | | Choice | 0 … 1 | | Elements to choose from:- hl7:subject containing template 1.3.6.1.4.1.19376.1.3.3.1.2.1 Non-Human Subject (DYNAMIC)
- hl7:subject containing template 1.3.6.1.4.1.19376.1.3.3.1.3.1 Human Patient with Non-Human Subject (DYNAMIC)
|  |  | hl7:subject
|
| | 0 … 1 | | subject in case of a non-human subject
Contains 1.3.6.1.4.1.19376.1.3.3.1.2.1 Non-Human Subject (DYNAMIC) | (Lab...ion) |  |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.2.1']] |
| | | cs | 0 … 1 | F | SBJ |  |  | hl7:subject
|
| | 0 … 1 | | subject in case of a non-human subject
Contains 1.3.6.1.4.1.19376.1.3.3.1.3.1 Human Patient with Non-Human Subject (DYNAMIC) | (Lab...ion) |  |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.3.1']] |
| | | cs | 0 … 1 | F | SBJ |  | hl7:performer
|
| | 0 … * | | performer participation. Performer to supersede those recorded at higher level.
Contains 1.3.6.1.4.1.19376.1.3.3.1.7 Laboratory Performer (DYNAMIC) | (Lab...ion) |  | where [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.3.1.7']] |
| |  |  | @typeCode
|
| cs | 0 … 1 | F | PRF |  | hl7:author
|
| | 0 … * | R | author participation used to supersede the authors of higher level.
Contains 1.3.6.1.4.1.19376.1.3.10.9.17 Author with Name, Addr, Telecom (DYNAMIC) | (Lab...ion) |  |  | @typeCode
|
| cs | 0 … 1 | F | AUT |  | hl7:participant
|
| | 0 … 1 | | Other participants "AUTHEN", "RESP", and "DEV". - AUTHEN: The verifier of the battery organizer. In the case where a laboratory report has multiple verifiers. Each verifier is attached to the subset of batteries/observations he or she verified, by the means of a <participant> element.
| (Lab...ion) |  | where [@typeCode='AUTHEN'] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | AUTHEN |  | hl7:participant
|
| | 0 … 1 | | Other participants "AUTHEN", "RESP", and "DEV". - RESP: The person responsible for the provision of the battery. In the case where this battery is subcontracted to an external laboratory, this external laboratory (with its address and telcom) and the actual performer is represented by a <performer> element, whereas the Director of this subcontractor laboratory is carried by the <participant> element
being attached to the same level as the <performer> element.
| (Lab...ion) |  | where [@typeCode='RESP'] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | RESP |  | hl7:participant
|
| | 0 … 1 | | Other participants "AUTHEN", "RESP", and "DEV". - DEV: A device or equipment, which was used to produce this battery of results (e.g. an analyzer).
| (Lab...ion) |  | where [@typeCode='DEV'] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | DEV |  | hl7:entryRelationship
|
| | 0 … * | R | Comment on this Observation
Contains 1.3.6.1.4.1.19376.1.5.3.1.4.2 IHE Comment Entry (DYNAMIC) | (Lab...ion) |  | where [hl7:act [hl7:templateId [@root='2.16.840.1.113883.10.20.1.40'] and hl7:templateId [@root='1.3.6.1.4.1.19376.1.5.3.1.4.2']]] |
| |  | hl7:entryRelationship
|
| | 0 … * | R | Specimen Collection
Contains 1.3.6.1.4.1.19376.1.3.1.2 Specimen Collection (DYNAMIC) | (Lab...ion) |  | where [hl7:procedure [hl7:templateId [@root='1.3.6.1.4.1.19376.1.3.1.2']]] |
| |  | hl7:entryRelationship
|
| | 0 … * | R | Previous observations obtained for the same patient, test, same method, same unit (1) | (Lab...ion) |  | where [@typeCode='REFR'] |
| |  |  | @typeCode
|
| cs | 1 … 1 | F | REFR |  |  | hl7:observation
|
| | 1 … 1 | R | | (Lab...ion) | | cs | 1 … 1 | F | OBS | | cs | 1 … 1 | F | EVN | | | 1 … 1 | R | | (Lab...ion) | | | 1 … 1 | R | | (Lab...ion) | | | 1 … 1 | F | completed | | | 1 … 1 | R | | (Lab...ion) | | | 1 … 1 | R | | (Lab...ion) |  | hl7:referenceRange
|
| | 0 … 1 | R | Reference range for the current test result | (Lab...ion) |  |  | @typeCode
|
| cs | 0 … 1 | F | REFV |  |  | hl7:observationRange
|
| | 1 … 1 | M | | (Lab...ion) | | cs | 0 … 1 | F | OBS | | cs | 0 … 1 | F | EVN.CRT | | | 0 … 1 | |  interval (IVL) representation | (Lab...ion) | | CE | 1 … 1 | R | | (Lab...ion) | | CONF | 0 … 1 | F | N | | 0 … 1 | F | 2.16.840.1.113883.5.83 (Observation Interpretation) | | | 0 … * | | Extension to CDA Clinical statement | (Lab...ion) | | cs | 1 … 1 | F | PRCN | | | 1 … 1 | R | | (Lab...ion) | | cs | 1 … 1 | F | COND | | cs | 1 … 1 | F | EVN | | CE | 1 … 1 | R | Code of the criterion (e.g. age, sex) | (Lab...ion) | | | 1 … 1 | R |  Value of the criterion | (Lab...ion) |
|
Human Patient (recordTarget) Child element - 1.3.6.1.4.1.19376.1.3.3.1.1
| Id | 1.3.6.1.4.1.19376.1.3.3.1.1 ref (from repository: XDLAB-) | Effective Date | valid from 2017‑06‑07 |
|---|
| Status |  Active Active | Version Label | 2017 |
|---|
| Name | Humanpatientrecordtarget | Display Name | Human Patient |
|---|
| Description | ClinicalDocument/recordTarget SHALL be present and SHALL conform to the Human Patient, Non-Human Subject or Human Patient with Non-Human Subject templates defined below. There are three varieties of laboratory reports: - Human (patient): The document reports laboratory observations produced on specimens collected exclusively from the patient.
- Non-Human Subject: The document reports laboratory observations produced on specimens collected from a non-human material (e.g. water, milk…) or living subject (e.g. animal).
- Human (patient) paired with Non-Human Subject: The document reports laboratory observations produced on a non-human specimen with a relationship to a human patient, (e.g. peanut butter eaten by a patient, a ferret that bit a patient).
Human Patient In accordance with the HL7 CDA R2 standard and further constrained by this specification, XD-LAB requires the presence of name, addr and telecom for all entities in the document including the human patient. Additionally, the following SHALL be present. - An optional templateId with root="1.3.6.1.4.1.19376.1.3.3.1.1" - The templateId element identifies this recordTarget as a human patient.
- id - The patientRole/id SHALL be present.
- administrativeGenderCode - The patientRole/patient/administrativeGenderCode SHALL be present.
- birthTime - The patientRole/patient/birthTime SHALL be present.
|
|
| Classification | CDA Header Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 1 template, Uses 0 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.1.1 | Include |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.101 (2005‑09‑07) |
|---|
| Example | | Human Patient Example a | <recordTarget typeCode="RCT">
<patientRole classCode="PAT">
<id extension="sw54321" root="1.3.6.1.4.1.19376.1.3.4"/> <addr>
<streetAddressLine>1313 Mockingbird Lane</streetAddressLine> <city>Janesville</city> <state>WI</state> <postalCode>53545</postalCode> <country>USA</country> </addr> <telecom value="tel: 608-555-5555"/> <patient classCode="PSN">
<name>
<family>Winters</family> <given>Shelly</given> </name> <administrativeGenderCode code="F"/> <birthTime value="19401213000000.0000-0500"/> </patient> </patientRole></recordTarget> |
|
| Example | | Human Patient Example b: In the event a unit of information about the patient is not known or has been de-identified, the use of nullFlavor is appropriate | <recordTarget typeCode="RCT">
<patientRole classCode="PAT">
<id extension="sw54321" root="1.3.6.1.4.1.19376.1.3.4"/> <addr>
<streetAddressLine nullFlavor="MSK"/> <!-- masked value -->
<city nullFlavor="MSK"/> <!-- masked value -->
<state nullFlavor="MSK"/> <!-- masked value -->
<postalCode>53545</postalCode> <country>USA</country> </addr> <telecom nullFlavor="UNK"/> <!-- unknown value -->
<patient classCode="PSN">
<name nullFlavor="MSK"/> <!-- masked value -->
<administrativeGenderCode code="F"/> <birthTime value="19401213000000.0000-0500"/> </patient> </patientRole></recordTarget> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| | 1 … * | R | | (Hum...get) |  | hl7:patientRole
|
| | 1 … 1 | R | | (Hum...get) |  |  | hl7:id
|
| II | 1 … * | R | | (Hum...get) |  |  | hl7:addr
|
| AD | 1 … * | | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom. | (Hum...get) |  |  | hl7:telecom
|
| TEL | 1 … * | | All persons (including the patient) and organizations mentioned in the document SHALL provide elements name, addr and telecom. | (Hum...get) |  |  | hl7:patient
|
| | 1 … 1 | | | (Hum...get) | | II | 0 … 1 | | | (Hum...get) | | PN | 1 … * | | | (Hum...get) |  |  |  | hl7:administrativeGenderCode
|
| CE | 1 … 1 | R | | (Hum...get) | | TS | 1 … 1 | R | | (Hum...get) |
|
Information Recipient Child element - 1.3.6.1.4.1.19376.1.3.3.1.4
| Id | 1.3.6.1.4.1.19376.1.3.3.1.4 | Effective Date | valid from 2016‑07‑09 15:17:01 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | CDAinformationRecipientIHE | Display Name | CDA informationRecipient IHE |
|---|
| Description | Template CDA informationRecipient (prototype, directly derived from POCD_RM000040 MIF) |
|---|
| Classification | CDA Header Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 1 template, Uses 2 templates | | Used by Template id | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.8.1.1.1 | |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | Uses Template id | as | Name | Version |
|---|
| 2.16.840.1.113883.10.12.152 | Containment |  CDA Person CDA Person | DYNAMIC | | 2.16.840.1.113883.10.12.151 | Containment |  CDA Organization CDA Organization | DYNAMIC |
|
|
|---|
| Relationship | Adaptation: template 2.16.840.1.113883.10.12.105 (2005‑09‑07) |
|---|
| Item | DT | Card | Conf | Description | Label |
|---|
hl7:informationRecipient
|
| | 0 … * | | | (CDAinformationRecipientIHE) |  | @typeCode
|
| cs | 1 … 1 | R | | | | CONF | | The value of @typeCode shall be drawn from value set 2.16.840.1.113883.1.11.19366 x_InformationRecipient (DYNAMIC) |
|  | hl7:intendedRecipient
|
| | 1 … 1 | R | | (CDAinformationRecipientIHE) |  |  | @classCode
|
| cs | 1 … 1 | R | | | | CONF | | The value of @classCode shall be drawn from value set 2.16.840.1.113883.1.11.16772 x_InformationRecipientRole (DYNAMIC) |
|  |  | hl7:id
|
| II | 0 … * | R | | (CDAinformationRecipientIHE) |  |  | hl7:addr
|
| AD | 0 … * | | | (CDAinformationRecipientIHE) |  |  | hl7:telecom
|
| TEL | 0 … * | | | (CDAinformationRecipientIHE) |  |  | hl7:informationRecipient
|
| | 0 … 1 | | Contains 2.16.840.1.113883.10.12.152 CDA Person (DYNAMIC) | (CDAinformationRecipientIHE) |  |  | hl7:receivedOrganization
|
| | 0 … 1 | | Contains 2.16.840.1.113883.10.12.151 CDA Organization (DYNAMIC) | (CDAinformationRecipientIHE) |
|
Ordering Provider Child element - 1.3.6.1.4.1.19376.1.3.3.1.6
Laboratory Performer Child element - 1.3.6.1.4.1.19376.1.3.3.1.7
| Id | 1.3.6.1.4.1.19376.1.3.3.1.7 ref (from repository: XDLAB-) | Effective Date | valid from 2008‑08‑08 |
|---|
| Status |  Active Active | Version Label | 2017 |
|---|
| Name | LaboratoryPerformer | Display Name | Laboratory Performer |
|---|
| Description | Laboratory Performers SHALL be recorded as performers in the CDA Header as well as in the structured body as demonstrated in the figure below. Documentation of laboratory performers MAY be done in multiple levels of the document to reflect performance scope. In the case where there is a single Laboratory Performer, this entity SHALL be documented in CDA header as shown. In the case where multiple Laboratory Performers participated in the lab testing process, they SHALL be documented in the structuredBody at the observation level. These performers SHALL conform to the General Constraints found in HL7 CRS: 2.1.1 with respect to the requirements for name, address, telephone numbers and other contact information. This module is out of
the XDS-Lab Specification and documented there in a <performer> element. This module does not change requirements stated in that specification but is structured as a module to ensure consistent content representation and re-usability. The figure below shows how the information for this element is coded, and further constraints are provided in the following sections. |
|
| Context | Sibling nodes of template element with id 1.3.6.1.4.1.19376.1.3.3.1.7 |
|---|
| Classification | CDA Header Level Template
CDA Entry Level Template |
|---|
| Open/Closed | Open (other than defined elements are allowed) |
|---|
| Used by / Uses | | Used by 0 transactions and 21 templates, Uses 0 templates | | Used by | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.1.6 | Containment |  Laboratory Observation (2017) Laboratory Observation (2017) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.3.6 |  |  Problem Organizer (2.0) Problem Organizer (2.0) | 2015‑08‑13 10:24:55 | | 1.3.6.1.4.1.19376.1.3.10.3.1 |  |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report Content Module (2.0) Anatomic Pathology Structured Report Content Module (2.0) | 2014‑05‑13 11:57:57 | | 1.3.6.1.4.1.19376.1.8.1.2.1 |  |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 |  |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 |  |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 |  |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.8.1.2.5 |  |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | 1.3.6.1.4.1.19376.1.3.10.4.1 | Containment |  Specimen Procedure Step (2.0) Specimen Procedure Step (2.0) | 2016‑07‑08 13:20:59 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.3.10.4.1 |  |  Specimen Procedure Step (1.0) Specimen Procedure Step (1.0) | 2014‑07‑29 16:02:02 | |  Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check Circular reference found with 1.3.6.1.4.1.19376.1.3.10.4.1, please check | | 1.3.6.1.4.1.19376.1.8.1.2.6 |  |  Procedure Steps Section (2.0) Procedure Steps Section (2.0) | 2014‑05‑13 19:33:12 | | 1.3.6.1.4.1.19376.1.3.10.4.2 | Containment |  TNM Stage Observation (2.0) TNM Stage Observation (2.0) | 2014‑05‑13 15:45:13 | | 1.3.6.1.4.1.19376.1.3.10.4.3 | Containment |  ICD-O-3 Typing and Grading Observation (2.0) ICD-O-3 Typing and Grading Observation (2.0) | 2015‑11‑29 17:03:18 | | 1.3.6.1.4.1.19376.1.3.10.4.4 | Containment |  Assessment Scales Observation for Scoring Systems APSR2 (2.0) Assessment Scales Observation for Scoring Systems APSR2 (2.0) | 2016‑10‑06 13:12:00 | | 1.3.6.1.4.1.19376.1.3.10.9.22 | Containment |  CDA Supply Container APSR2 (2.0) CDA Supply Container APSR2 (2.0) | 2016‑07‑29 12:09:33 | | 1.3.6.1.4.1.19376.1.3.10.9.25 | Containment |  TNM T-Observation (2.0) TNM T-Observation (2.0) | 2014‑12‑02 15:54:17 | | 1.3.6.1.4.1.19376.1.3.10.9.43 | Containment |  Assessment Scoring Item APSR 2.0 (2.0) Assessment Scoring Item APSR 2.0 (2.0) | 2016‑10‑11 10:26:17 | | 1.3.6.1.4.1.19376.1.3.10.9.42 |  |  Assessment Scoring System APSR 2.0 (2.0) Assessment Scoring System APSR 2.0 (2.0) | 2016‑10‑10 11:33:36 | | 1.3.6.1.4.1.19376.1.8.1.4.10 | Containment |  Embedded Image IHE Embedded Image IHE | 2016‑06‑22 16:08:01 | | 1.3.6.1.4.1.19376.1.8.1.4.9 |  |  AP Observation Entry (2.0) AP Observation Entry (2.0) | 2014‑05‑14 17:09:54 | |  Circular reference found with 1.3.6.1.4.1.19376.1.8.1.4.9, please check Circular reference found with 1.3.6.1.4.1.19376.1.8.1.4.9, please check |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.320 (2005‑09‑07) |
|---|
| Example | | Laboratory Performer at Service Event Level (Header) | <serviceEvent>
<!-- .. -->
<performer typeCode="PRF">
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.7"/> <assignedEntity>
<id extension="kd83736" root="1.3.6.1.4.1.19376.1.3.4"/> <addr>
<streetAddressLine>7000 Hosptial Drive</streetAddressLine> <city>Chicago</city> <state>IL</state> <postalCode>60622</postalCode> <country>USA</country> </addr> <telecom value="tel:312-555-5555"/> <assignedPerson>
<name>
<family>Dawson</family> <given>Kim</given> <prefix>Dr.</prefix> </name> </assignedPerson> <representedOrganization>
<id extension="9875" root="1.3.6.1.4.1.19376.1.3.4"/> <name>PRF Hospital Laboratory</name> <telecom value="tel:312-555-5555"/> <addr>
<streetAddressLine>7000 Hosptial Drive</streetAddressLine> <city>Chicago</city> <state>IL</state> <postalCode>60622</postalCode> <country>USA</country> </addr> </representedOrganization> </assignedEntity> </performer></serviceEvent> |
|
| Example | | Laboratory Performer at Observation Level (Body) | <observation>
<!-- .. -->
<performer typeCode="PRF">
<templateId root="1.3.6.1.4.1.19376.1.3.3.1.7"/> <assignedEntity>
<id extension="rm83747" root="1.3.6.1.4.1.19376.1.3.4"/> <addr>
<streetAddressLine>7000 Hosptial Drive</streetAddressLine> <city>Chicago</city> <state>IL</state> <postalCode>60622</postalCode> <country>USA</country> </addr> <telecom value="tel:312-555-5555"/> <assignedPerson>
<name>
<family>Trenton</family> <given>Douglas</given> <prefix>Dr.</prefix> </name> </assignedPerson> <representedOrganization>
<id extension="rm83747" root="1.3.6.1.4.1.19376.1.3.4"/> <name>PRF Hospital Laboratory</name> <telecom value="tel:312-555-5555"/> <addr>
<streetAddressLine>7000 Hosptial Drive</streetAddressLine> <city>Chicago</city> <state>IL</state> <postalCode>60622</postalCode> <country>USA</country> </addr> </representedOrganization> </assignedEntity> </performer></observation> |
|
| Item | DT | Card | Conf | Description | Label |
|---|
| cs | 0 … 1 | F | PRF | | II | 1 … 1 | M | | (Lab...mer) |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.3.3.1.7 | | IVL_TS | 1 … 1 | R | The element SHALL be present and SHALL represent the point in time of laboratory performance. | (Lab...mer) | | | 1 … 1 | | | (Lab...mer) |  | @classCode
|
| cs | 0 … 1 | F | ASSIGNED |  | hl7:id
|
| II | 1 … * | R | Unique identifier of this person/organization (lab performer) in the affinity domain SHALL be present. | (Lab...mer) |  | hl7:code
|
| CE | 0 … 1 | R | | (Lab...mer) |  | hl7:addr
|
| AD | 1 … * | R | The address of this person/organization (lab performer) SHALL be present. | (Lab...mer) |  | hl7:telecom
|
| TEL | 1 … * | R | The telecom of this person/organization (lab performer) SHALL be present. | (Lab...mer) |  | hl7:assignedPerson
|
| | 0 … 1 | R | | (Lab...mer) |  | hl7:representedOrganization
|
| | 0 … 1 | R | | (Lab...mer) | | | Schematron assert | role |  error error | | | | test | hl7:assignedPerson/hl7:name or hl7:representedOrganization/hl7:name | | | | Message | Either an <assignedPerson> or a <representedOrganization> SHALL be present. In either case, the <name> sub-element SHALL be present. | |
|
Comment Child element - 1.3.6.1.4.1.19376.1.5.3.1.4.2
| Id | 1.3.6.1.4.1.19376.1.5.3.1.4.2 | Effective Date | valid from 2016‑06‑23 14:21:01 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | AnnotationCommentIHE | Display Name | Annotation Comment (IHE) |
|---|
| Description | This entry is used for Annotation Comments in APSR 2.0 |
|---|
| Context | Parent nodes of template element with id 1.3.6.1.4.1.19376.1.5.3.1.4.2 |
|---|
| Classification | CDA Entry Level Template |
|---|
| Open/Closed | Closed (only defined elements are allowed) |
|---|
| Used by / Uses | | Used by 12 templates, Uses 0 templates | | Used by Template id | as | Name | Version |
|---|
| 1.3.6.1.4.1.19376.1.3.1.6 | Containment |  Laboratory Observation (2016) Laboratory Observation (2016) | 2016‑07‑05 | | 1.3.6.1.4.1.19376.1.3.1.6 |  |  Laboratory Observation (2008) Laboratory Observation (2008) | 2008‑08‑08 | | 1.3.6.1.4.1.19376.1.8.1.3.6 |  |  Problem Organizer (2.0) Problem Organizer (2.0) | 2015‑08‑13 10:24:55 | | 1.3.6.1.4.1.19376.1.3.10.3.1 |  |  Additional Specified Observation Section (2.0) Additional Specified Observation Section (2.0) | 2016‑11‑13 14:28:08 | | 1.3.6.1.4.1.19376.1.8.1.1.1 |  |  Anatomic Pathology Structured Report (2.0) Anatomic Pathology Structured Report (2.0) | 2014‑05‑13 11:57:57 | | 1.3.6.1.4.1.19376.1.8.1.2.1 |  |  Clinical Information Section (2.0) Clinical Information Section (2.0) | 2014‑05‑13 14:38:08 | | 1.3.6.1.4.1.19376.1.8.1.2.2 |  |  Intraoperative Observation Section (2.0) Intraoperative Observation Section (2.0) | 2014‑05‑13 19:29:16 | | 1.3.6.1.4.1.19376.1.8.1.2.3 |  |  Macroscopic Observation Section (2.0) Macroscopic Observation Section (2.0) | 2014‑05‑13 11:57:09 | | 1.3.6.1.4.1.19376.1.8.1.2.4 |  |  Microscopic Observation Section (2.0) Microscopic Observation Section (2.0) | 2014‑05‑13 14:25:17 | | 1.3.6.1.4.1.19376.1.8.1.2.5 |  |  Diagnostic Conclusion Section (2.0) Diagnostic Conclusion Section (2.0) | 2014‑05‑13 19:31:26 | | 1.3.6.1.4.1.19376.1.8.1.4.10 | Containment |  Embedded Image IHE Embedded Image IHE | 2016‑06‑22 16:08:01 | | 1.3.6.1.4.1.19376.1.8.1.4.9 |  |  AP Observation Entry (2.0) AP Observation Entry (2.0) | 2014‑05‑14 17:09:54 |
|
|
|---|
| Relationship | Specialization: template 2.16.840.1.113883.10.12.303 (2005‑09‑07)
Specialization: template 2.16.840.1.113883.10.20.1.40 (DYNAMIC)
Specialization: template 1.3.6.1.4.1.19376.1.5.3.1.4.2 (2009‑12‑04)
Adaptation: template 1.2.276.0.76.10.4015 (2014‑11‑15) |
|---|
| Item | DT | Card | Conf | Description | Label |
|---|
| | | | The related statement is an event (moodCode='EVN') describing the act (classCode='ACT') of making an arbitrary comment or providing instruction on the related entry. | (AnnotationCommentIHE) |  | @classCode
|
| cs | 1 … 1 | F | ACT |  | @moodCode
|
| cs | 1 … 1 | F | EVN |  | hl7:templateId
|
| II | 1 … 1 | M | | (AnnotationCommentIHE) |  |  | @root
|
| uid | 1 … 1 | F | 1.3.6.1.4.1.19376.1.5.3.1.4.2 |  | hl7:templateId
|
| II | 1 … 1 | M | | (AnnotationCommentIHE) |  |  | @root
|
| uid | 1 … 1 | F | 2.16.840.1.113883.2.6.60.6.10.50 |  | hl7:code
|
| CD | 1 … 1 | M | The <code> element indicates that this is a comment and shall be recorded as shown above. The codeSystem and code attributes shall use the values specified above. | (AnnotationCommentIHE) |  |  | @code
|
| CONF | 1 … 1 | F | 48767-8 |  |  | @codeSystem
|
| 1 … 1 | F | 2.16.840.1.113883.6.1 (Logical Observation Identifier Names and Codes) |  |  | @displayName
|
| 1 … 1 | F | Annotation Comment |  |  | @codeSystemName
|
| 1 … 1 | F | LOINC |  | hl7:text
|
| ED | 1 … 1 | M | The <text> element provides a way to represent the <reference> to the text of the comment in the narrative portion of the document. For CDA, this SHALL be represented as a <reference> element that points to the narrative text section of the CDA. The comment itself is not the act being coded, so it appears in the <text> of the <observation>, not as part of the <code>. For HL7 Version 3 Messages, the <text> element SHALL contain the full narrative text. | (AnnotationCommentIHE) |  | hl7:statusCode
|
| CS | 1 … 1 | M | The code attribute of <statusCode> for all comments must be completed. | (AnnotationCommentIHE) | | | CONF | |
|
Appendix A - Value Sets
A. APSR Value Sets
There are both extensional and intensional value sets specifically used for APSR.
A.1 Generic AP Observation Codes 1.3.6.1.4.1.19376.1.3.11.11
| Id | 1.3.6.1.4.1.19376.1.3.11.11 | Effective Date | valid from 2015‑11‑22 12:20:34 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | AP-Observation | Display Name | AP Observation |
|---|
| Description | Generic codes for AP Observations of all kind, including results of ancillary techniques (immunohistochemistry, in-situ-hybridization, other molecular methods) |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
A valid code from one of the 12 code systems:| Code System Name | Code System Id | Code System Version | Flexibility | | Logical Observation Identifier Names and Codes | 2.16.840.1.113883.6.1 | | DYNAMIC | | SNOMED Clinical Terms | 2.16.840.1.113883.6.96 | | DYNAMIC | | International Classification of Diseases for Oncology, version 3. | 2.16.840.1.113883.6.43.1 | | DYNAMIC | | ICD10 | 2.16.840.1.113883.6.3 | | DYNAMIC | | TNM Edition8 UICC | 2.16.840.1.113883.15.16 | | DYNAMIC | | TNM Edition7 UICC | 2.16.840.1.113883.15.6 | | DYNAMIC | | TNM Edition6 UICC | 2.16.840.1.113883.15.7 | | DYNAMIC | | TNM Edition5 UICC | 2.16.840.1.113883.15.8 | | DYNAMIC | | Unified Code for Units of Measure | 2.16.840.1.113883.6.8 | | DYNAMIC | | Online Mendelian Inheritance in Man 1993 | 2.16.840.1.113883.6.174 | | DYNAMIC | | DKG Coding scheme | 1.2.276.0.76.3.1.131.1.5.1 | | DYNAMIC | | localization codes for distant metastases | 1.2.276.0.76.5.401 | | DYNAMIC |
|
A.2 Problem type 1.3.6.1.4.1.19376.1.3.11.7
| Id | 1.3.6.1.4.1.19376.1.3.11.7 | Effective Date | valid from 2016‑07‑11 09:52:19 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | ProblemType | Display Name | Problem Type |
|---|
| Description | Values for the problem under investigation |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
A valid code from the code system:| Code System Name | Code System Id | Code System Version | | ICD10 | 2.16.840.1.113883.6.3 | |
|
| Or one of the following: |
| 4 Source Code Systems | 2.16.840.1.113883.6.3 - ICD10 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 2.16.840.1.113883.5.6 - ActClass 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 115597007 | Description of specimen character (procedure) | SNOMED Clinical Terms | | 0‑L | 128462008 | Secondary malignant neoplastic disease (disorder) | SNOMED Clinical Terms | | 0‑L | 367651003 | Malignant neoplasm of primary, secondary, or uncertain origin (morphologic abnormality) | SNOMED Clinical Terms | | 0‑L | SPCOBS | specimen observation | ActClass | | 0‑L | 3898006 | Neoplasm, benign (morphologic abnormality) | SNOMED Clinical Terms | | 0‑L | 109355002 | Carcinoma in situ (disorder) | SNOMED Clinical Terms | | 0‑L | 23583003 | Inflammation (morphologic abnormality) | SNOMED Clinical Terms | | 0‑L | 33359002 | Degeneration (morphologic abnormality) | SNOMED Clinical Terms | | 0‑L | 49601007 | Disorder of cardiovascular system (disorder) | SNOMED Clinical Terms | | 0‑L | ASKU | asked but unknown | Null Flavor |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.3 Container Entity Class Type 1.3.6.1.4.1.19376.1.3.11.8
| Id | 1.3.6.1.4.1.19376.1.3.11.8 | Effective Date | valid from 2016‑09‑08 11:55:11 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | ContainerEntityClassTypePaLM | Display Name | ContainerEntityClassTypePaLM |
|---|
| Description | Types of Containers and their additives in Anatomic Pathology |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 1.2.840.10008.2.16.4 - DICOM Controlled Terminology |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 434746001 | Specimen vial (physical object) | SNOMED Clinical Terms | | 0‑L | 434464009 | Tissue cassette (physical object) | SNOMED Clinical Terms | | 0‑L | 434708008 | Tissue cassette for microarray (physical object) | SNOMED Clinical Terms | | 0‑L | 433466003 | Microscope slide (physical object) | SNOMED Clinical Terms | | 0‑S | 433453003 | Specimen container component (physical object) | SNOMED Clinical Terms | | 1‑S | 430863003 | Tissue embedding medium (substance) | SNOMED Clinical Terms | | 2‑L | 311731000 | Paraffin wax (substance) | SNOMED Clinical Terms | | 2‑L | 433469005 | Frozen section embedding medium (substance) | SNOMED Clinical Terms | | 2‑L | 61088005 | Plastic (substance) | SNOMED Clinical Terms | | 2‑L | 10249006 | Agar (substance) | SNOMED Clinical Terms | | 2‑L | 65345002 | Epoxy resin (substance) | SNOMED Clinical Terms | | 2‑L | 427811002 | Polymethyl methacrylate (substance) | SNOMED Clinical Terms | | 1‑L | 433472003 | Microscope slide coverslip (physical object) | SNOMED Clinical Terms | | 1‑L | 430862008 | Microscope slide mounting medium (substance) | SNOMED Clinical Terms | | 0‑S | CID8101 | Container type | DICOM Controlled Terminology | | 1‑L | A-01024 | Specimen vial | DICOM Controlled Terminology | | 1‑L | A-0101B | Microscope slide | DICOM Controlled Terminology | | 1‑L | A-01023 | Specimen container | DICOM Controlled Terminology | | 1‑L | A-01021 | Electron microscopy grid | DICOM Controlled Terminology | | 1‑L | A-01025 | Specimen well | DICOM Controlled Terminology | | 0‑S | CID8114 | Fixatives | DICOM Controlled Terminology | | 0‑S | CID8102 | Container Component Types | DICOM Controlled Terminology | | 0‑S | CID8115 | Specimen Embedding Media | DICOM Controlled Terminology |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.4 Procedure (target) site 1.3.6.1.4.1.19376.1.3.11.9
| Id | 1.3.6.1.4.1.19376.1.3.11.9 | Effective Date | valid from 2016‑06‑01 18:46:15 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | ProcedureSite | Display Name | Procedure Site |
|---|
| Description | Target sites of specimen processing procedures |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 1.2.840.10008.2.16.4 - DICOM Controlled Terminology |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | 309050000 | Body substance sample | SNOMED Clinical Terms | all children of "Body substance sample" | | 0‑L | 309051001 | Body fluid sample | SNOMED Clinical Terms | | | 0‑S | 430856003 | Tissue section (specimen) | SNOMED Clinical Terms | | | 1‑L | 441652008 | Formalin-fixed paraffin-embedded tissue specimen | SNOMED Clinical Terms | | | 1‑L | 16214131000119104 | Frozen tissue section sample (specimen) | SNOMED Clinical Terms | | | 0‑S | CID8103 | Anatomic Pathology Specimen Types | DICOM Controlled Terminology | | | 1‑L | G-843A | Gross specimen | DICOM Controlled Terminology | | | 1‑L | G-843B | Core sample of tissue block | DICOM Controlled Terminology | | | 1‑L | G-843C | Tissue spot | DICOM Controlled Terminology | | | 1‑L | G-803C | Smear sample | DICOM Controlled Terminology | | | 1‑L | T-1A404 | Touch preparation cytologic material | DICOM Controlled Terminology | | | 1‑L | T-1A403 | Liquid based cytologic material | DICOM Controlled Terminology | | | 1‑L | G-8003 | Aspirate | DICOM Controlled Terminology | | | 0‑L | 48469005 | Cytologic material (specimen) | SNOMED Clinical Terms | |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.5 Specimen collection and processing 1.3.6.1.4.1.19376.1.3.11.10
| Id | 1.3.6.1.4.1.19376.1.3.11.10 | Effective Date | valid from 2016‑05‑31 15:09:59 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | SpecimenCollectionAndProcessing | Display Name | Specimen Collection and Processing |
|---|
| Description | Mixed Value set for Specimen collection and Processing |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 5 Source Code Systems | 2.16.840.1.113883.5.6 - ActClass 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 1.2.276.0.76.5.464 1.2.840.10008.2.16.4 - DICOM Controlled Terminology 2.16.840.1.113883.6.4 - International Classification of Diseases, 10th Revision, Procedure Coding System (ICD-10-PCS) |
|---|
| Level/ Type | Code | Display Name | Code System | Designations |
|---|
| 0‑S | SPECCOLLECT | Specimen collection | ActClass | | | 1‑S | 17636008 | Specimen collection (procedure) | SNOMED Clinical Terms | | | 2‑L | 48635004 | Fine needle biopsy (procedure) | SNOMED Clinical Terms | | | 2‑S | 86273004 | Biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 439336003 | Brush biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 53767003 | Endoscopic biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 8889005 | Excisional biopsy (procedure) | SNOMED Clinical Terms | | | 3‑S | 277590007 | Imaging guided biopsy (procedure) | SNOMED Clinical Terms | | | 4‑L | 442787002 | Fine needle aspiration biopsy using imaging guidance (procedure) | SNOMED Clinical Terms | | | 4‑L | 277592004 | Magnetic resonance imaging guided biopsy (procedure) | SNOMED Clinical Terms | | | 4‑L | 277667006 | Ultrasound guided biopsy (procedure) | SNOMED Clinical Terms | | | 4‑L | 277666002 | X-ray guided biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 45681003 | Staging operation involving incision, exploration and biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 70871006 | Incisional biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 129249002 | Needle biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 119283008 | Open biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 68660007 | Punch biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 72342005 | Shave biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 274330002 | Surgical biopsy (procedure) | SNOMED Clinical Terms | | | 3‑L | 50590007 | Trephine biopsy (procedure) | SNOMED Clinical Terms | | | 2‑S | 65801008 | Excision (procedure) | SNOMED Clinical Terms | | | 3‑L | 81723002 | Amputation (procedure) | SNOMED Clinical Terms | | | 3‑L | 86743009 | Local excision (procedure) | SNOMED Clinical Terms | | | 3‑L | 125571002 | Lobectomy (procedure) | SNOMED Clinical Terms | | | 3‑L | 26007001 | Hemi-excision (procedure) | SNOMED Clinical Terms | | | 3‑L | 38829003 | Partial excision (procedure) | SNOMED Clinical Terms | | | 3‑L | 89540006 | Segmental excision and ligation (procedure) | SNOMED Clinical Terms | | | 3‑L | 65854006 | Reexcision (procedure) | SNOMED Clinical Terms | | | 3‑S | 62141006 | Radical excision (procedure) | SNOMED Clinical Terms | | | 4‑L | 88088007 | Radical excision with lymph node dissection (procedure) | SNOMED Clinical Terms | | | 3‑L | 79095000 | Complete excision of organ (procedure) | SNOMED Clinical Terms | | | 3‑L | 370612006 | Excision of neoplasm (procedure) | SNOMED Clinical Terms | | | 3‑L | 20418004 | Wedge resection (procedure) | SNOMED Clinical Terms | | | 2‑L | 56757003 | Scraping (procedure) | SNOMED Clinical Terms | | | 1‑L | 1-40...1-49 | Biopsies without incision | 1.2.276.0.76.5.464 | | | 1‑L | 1-50...1-58 | Biopsies with incision | 1.2.276.0.76.5.464 | | | 1‑L | 5-01...5-09 | Surgical measures | 1.2.276.0.76.5.464 | | | 1‑L | 1-10...1-99 | Diagnostic measures | 1.2.276.0.76.5.464 | | | 1‑S | CID8109 | all children of Specimen Collection Procedure | DICOM Controlled Terminology | | | 1‑S | 00...0Y | all children of 0 | International Classification of Diseases, 10th Revision, Procedure Coding System (ICD-10-PCS) | | | 0‑S | SPCTRT | Specimen treatment | ActClass | | | 1‑S | CID8110 | all children of Specimen Sampling Procedures | DICOM Controlled Terminology | | | 1‑S | CID8111 | all children of Specimen Preparation Procedure | DICOM Controlled Terminology | | | 1‑S | CID8112 | all children of Specimen Stains | DICOM Controlled Terminology | | | 1‑S | CID8113 | all children of Specimen Preparation Steps | DICOM Controlled Terminology | | | 1‑S | CID8114 | all children of Specimen Fixatives | DICOM Controlled Terminology | | | 1‑S | 9265001 | Specimen processing (procedure) | SNOMED Clinical Terms | | | 2‑L | 430854000 | Touch preparation of specimen (procedure) | SNOMED Clinical Terms | | | 2‑L | 448938001 | Preparation of smear (procedure) | SNOMED Clinical Terms | | | 2‑L | 77896007 | Cytopathology procedure, cell block and smear preparation (procedure) | SNOMED Clinical Terms | | | 2‑L | 4064007 | Specimen aliquoting (procedure) | SNOMED Clinical Terms | | | 2‑L | 8378006 | Trimming (procedure) | SNOMED Clinical Terms | | | 2‑L | 73373003 | Specimen centrifugation (procedure) | SNOMED Clinical Terms | | | 2‑L | 27872000 | Specimen freezing (procedure) | SNOMED Clinical Terms | | | 2‑L | 104166004 | Nucleic acid molecular isolation or extraction method (procedure) | SNOMED Clinical Terms | | | 2‑S | 13283003 | Tissue processing technique (procedure) | SNOMED Clinical Terms | | | 3‑L | 27204007 | Tissue frozen section technique, complete (procedure) | SNOMED Clinical Terms | | | 3‑L | 40923002 | Tissue processing technique, routine, embed, cut and stain, per surgical specimen (procedure) | SNOMED Clinical Terms | | | 4‑L | 434472006 | Sectioning of tissue block (procedure) | SNOMED Clinical Terms | | | 4‑L | 434475008 | Step sectioning of tissue block (procedure) | SNOMED Clinical Terms | | | 4‑L | 434474007 | Surface recutting of tissue block (procedure) | SNOMED Clinical Terms | | | 4‑L | 434479002 | Core sampling of tissue block (procedure) | SNOMED Clinical Terms | | | 4‑L | 433454009 | Microdissection of tissue specimen using laser (procedure) | SNOMED Clinical Terms | | | 4‑L | 702941008 | Paraffin embedding (qualifier value) | SNOMED Clinical Terms | | | 2‑S | 127790008 | Staining method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406792001 | Acid phosphatase stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406901007 | Neutrophil alkaline phosphatase stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406902000 | New fuchsin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406903005 | Nile blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406904004 | Nitrazine yellow stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117229004 | Nonspecific esterase stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406905003 | Nuclear fast red stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406906002 | Orange G stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406907006 | Orange II stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406908001 | Page blue 83 stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406909009 | Page blue G-90 stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406910004 | Patent blue V sodium salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406911000 | Permethrin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406912007 | Phloxin B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406913002 | Ponceau 3R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406914008 | Ponceau S stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406915009 | Ponceau xylidine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406916005 | Pontamine sky blue 5BX stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406900008 | Naphthol yellow S stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406899003 | Naphthol green B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406898006 | Naphthalene black 12B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117306005 | Lawson-Van Gieson stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406875003 | Leishman stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406876002 | Light green SF stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406877006 | Lissamine fast red B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406878001 | Lissamine fast yellow stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406880007 | Lissamine rhodamine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 398507009 | Malachite green stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117218003 | Mallory bleach stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406890004 | Martius yellow stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406891000 | Meldola blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406892007 | Metanil yellow stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406893002 | Methyl blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406894008 | Methyl orange stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406895009 | Methyl red stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406896005 | Methylene blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406897001 | Methylene violet stain (Bernthsen) method (procedure) | SNOMED Clinical Terms | | | 3‑L | 37926009 | Microbial stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406917001 | Pontamine sky blue 6BX stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406918006 | Procion brilliant blue MRS stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117326009 | Quinacrine fluorescent stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406936006 | Sudan blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406937002 | Sunset yellow FCF stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117031001 | Supravital stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406938007 | Tartrazine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406939004 | Thioflavine T stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406940002 | Thionine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406941003 | Titan yellow stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406942005 | Tropaeolin O stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406943000 | Tropaeolin OO stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406944006 | Trypan blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406945007 | Vital new red stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 104249006 | Vital stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406946008 | Water soluble aniline blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406948009 | Water soluble anthracene brown stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406947004 | Water soluble nigrosine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406949001 | Waxoline blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406950001 | Xylene cyanol FF stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406934009 | Sudan black stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406935005 | Sudan black B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117030000 | Sudan IV stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406919003 | Romanowsky stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406920009 | Rose bengal stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406921008 | Rosolic acid sodium salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406922001 | Saffron stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117330007 | Schmorl stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117041003 | Silver stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406924000 | Sirius red F3B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406925004 | Solochrome azurine (BS) stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406926003 | Solochrome black 6B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406927007 | Solochrome cyanine R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406928002 | Solochrome dark blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406929005 | Soluble berlin blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406930000 | Spirit soluble aniline blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406931001 | Spirit soluble eosin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406932008 | Spore stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406933003 | Sudan II stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117029005 | Sudan III stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406874004 | Lacmoid stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406873005 | Kenacid blue R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406818007 | Bismark brown Y stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406819004 | Blue shade eosin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406820005 | Brazilin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406821009 | Brilliant cresyl blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406822002 | Brilliant crocein stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406823007 | Brilliant orange stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406824001 | Brilliant yellow stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 77281003 | Calcofluor white stain (procedure) | SNOMED Clinical Terms | | | 3‑L | 406825000 | Carminic acid stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406826004 | Carmoisine A stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406827008 | Celestine blue B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406828003 | Chloroacetate esterase stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406830001 | Chromotrope 2R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406831002 | Chrysoidine R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406832009 | Chrysoidine Y stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406833004 | Cochineal stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117055006 | Congo red stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406817002 | Bismark brown R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406816006 | Biebrich scarlet stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406815005 | Beta-glucuronidase stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406793006 | Albert's stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117036006 | Alcian blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406794000 | Alcohol soluble nigrosine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406795004 | Alizarin blue S stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406796003 | Alizarin cyanine green stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406797007 | Alizarin red S stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406798002 | Alizarin yellow GG stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406799005 | Alizarin yellow R stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406800009 | Auramine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406801008 | Azo black stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406802001 | Azocarmine G (GX) stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406803006 | Azophloxin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406804000 | Azorubin S stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406811001 | Azure A stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406812008 | Azure B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406813003 | Azure C stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406814009 | Benzo fast scarlet stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406835006 | Crystal ponceau stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406836007 | Curcumin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117298004 | Cytopathology staining method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406857004 | Flagellar stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406858009 | Fluorescent stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406863008 | Fuchsin acid stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 117047004 | Fuchsin basic stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 20543007 | Fungus stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406864002 | Gallocyanine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 422329009 | Hall's bilirubin stain (procedure) | SNOMED Clinical Terms | | | 3‑L | 406865001 | Hematein stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 108256005 | Hematology staining procedure (procedure) | SNOMED Clinical Terms | | | 3‑L | 406867009 | Immunofluorescent stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406868004 | Indigo carmine stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406869007 | Indophenol from naphthol stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406870008 | Insoluble berlin blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406871007 | Janus green B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406872000 | Jenner-Giemsa stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406856008 | Field's stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406855007 | Feulgen reaction stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406854006 | Fat red 7B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406837003 | Diamond black stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406838008 | Durazol red stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406839000 | Erie garnet stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406840003 | Eriochrome blue black SE stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406841004 | Erythrosin B stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406842006 | Erythrosin Y stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406843001 | Evans blue stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406844007 | Fast blue B salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406845008 | Fast blue BB salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406846009 | Fast blue RR salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406847000 | Fast garnet GBC salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406848005 | Fast green FCF stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406849002 | Fast red B salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406850002 | Fast red ITR stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406851003 | Fast red TR salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406852005 | Fast sulfon black F stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406853000 | Fast violet B salt stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 406979008 | Ziehl-Neelsen stain method (procedure) | SNOMED Clinical Terms | | | 3‑S | 104210008 | Hematoxylin and eosin stain method (procedure) | SNOMED Clinical Terms | | | 4‑L | 117239005 | Harris regressive hematoxylin and eosin stain method (procedure) | SNOMED Clinical Terms | | | 4‑L | 117240007 | Mayers progressive hematoxylin and eosin stain method (procedure) | SNOMED Clinical Terms | | | 3‑L | 80246006 | Special stain, blood or bone marrow, explain by report (procedure) | SNOMED Clinical Terms | | | 3‑L | 117617002 | Immunohistochemistry procedure (procedure) | SNOMED Clinical Terms | | | 3‑L | 384715000 | Nucleic acid hybridization procedure (procedure) | SNOMED Clinical Terms | |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.6 UICC/AJCC stage 1.3.6.1.4.1.19376.1.3.11.46
| Id | 1.3.6.1.4.1.19376.1.3.11.46 | Effective Date | valid from 2017‑07‑21 10:43:31 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICCstage-8ed | Display Name | UICC Stage |
|---|
| Description | UICC Stages
|
|---|
| 3 Source Code Systems | 2.16.840.1.113883.15.6 - TNM Edition7 UICC 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | okk | stage X | TNM Edition7 UICC | TX, N0, M0 | | 0‑S | 0 | stage 0 | TNM Edition7 UICC | Tis N0 M0 | | 1‑L | 0a | stage 0a | TNM Edition7 UICC | in C67 only | | 1‑L | 0is | stage 0is | TNM Edition7 UICC | in C67 only | | 0‑S | I | stage I | TNM Edition7 UICC | organ dependent | | 2‑L | IA1 | stage IA1 | TNM Edition7 UICC | organ dependent
| | 2‑L | IA2 | stage IA2 | TNM Edition7 UICC | organ dependent
| | 1‑S | IB | stage IB | TNM Edition7 UICC | organ dependent | | 2‑L | IB1 | stage IB1 | TNM Edition7 UICC | organ dependent
| | 2‑L | IB2 | stage IB2 | TNM Edition7 UICC | organ dependent
| | 1‑L | IC | stage IC | TNM Edition7 UICC | organ dependent
| | 1‑L | IS | stage IS | TNM Edition7 UICC | in C62 only | | 0‑S | II | stage II | TNM Edition7 UICC | organ dependent
| | 2‑L | IIA | stage IIA | TNM Edition7 UICC | organ dependent
| | 2‑L | IIA1 | stage IIA1 | TNM Edition7 UICC | organ dependent
| | 2‑L | IIA2 | stage IIA2 | TNM Edition7 UICC | organ dependent
| | 1‑L | IIB | stage IIB | TNM Edition7 UICC | organ dependent
| | 1‑L | IIC | stage IIC | TNM Edition7 UICC | organ dependent
| | 0‑S | III | stage III | TNM Edition7 UICC | organ dependent
| | 1‑L | IIIA | stage IIIA | TNM Edition7 UICC | organ dependent
| | 1‑L | IIIB | stage IIIB | TNM Edition7 UICC | organ dependent
| | 1‑L | IIIC | stage IIIC | TNM Edition7 UICC | organ dependent
| | 0‑S | IV | stage IV | TNM Edition7 UICC | each T, each N, M1 | | 1‑L | IVA | stage IVA | TNM Edition7 UICC | organ dependent
| | 1‑L | IVB | stage IVB | TNM Edition7 UICC | organ dependent
| | 1‑L | IVC | stage IVC | TNM Edition7 UICC | organ dependent
| | 0‑S | 0 | stage 0 | TNM Edition8 UICC | Tis N0 M0 | | 1‑L | 0a | stage 0a | TNM Edition8 UICC | in C67 only | | 1‑L | 0is | stage 0is | TNM Edition8 UICC | in C67 only | | 0‑S | I | stage I | TNM Edition8 UICC | organ dependent | | 1‑S | IA | stage IA | TNM Edition8 UICC | | | 2‑L | IA1 | stage IA1 | TNM Edition8 UICC | organ dependent
| | 2‑L | IA2 | stage IA2 | TNM Edition8 UICC | organ dependent
| | 2‑L | IA3 | stage IA3 | TNM Edition8 UICC | | | 1‑S | IB | stage IB | TNM Edition8 UICC | organ dependent | | 2‑L | IB1 | stage IB1 | TNM Edition8 UICC | organ dependent
| | 2‑L | IB2 | stage IB2 | TNM Edition8 UICC | organ dependent
| | 1‑L | IC | stage IC | TNM Edition8 UICC | organ dependent
| | 1‑L | IS | stage IS | TNM Edition8 UICC | in C62 only | | 0‑S | II | stage II | TNM Edition8 UICC | organ dependent
| | 1‑S | IIA | stage IIA | TNM Edition7 UICC | organ dependent
| | 2‑L | IIA1 | stage IIA1 | TNM Edition8 UICC | organ dependent
| | 2‑L | IIA2 | stage IIA2 | TNM Edition8 UICC | organ dependent
| | 1‑L | IIB | stage IIB | TNM Edition8 UICC | organ dependent
| | 1‑L | IIC | stage IIC | TNM Edition8 UICC | organ dependent
| | 0‑S | III | stage III | TNM Edition8 UICC | organ dependent
| | 1‑S | IIIA | stage IIIA | TNM Edition8 UICC | organ dependent
| | 2‑L | IIIA1 | stage IIIA1 | TNM Edition8 UICC | | | 2‑L | IIIA2 | stage IIIA2 | TNM Edition8 UICC | | | 1‑L | IIIB | stage IIIB | TNM Edition8 UICC | organ dependent
| | 1‑S | IIIC | stage IIIC | TNM Edition8 UICC | organ dependent
| | 2‑L | IIIC1 | stage IIIC1 | TNM Edition8 UICC | | | 2‑L | IIIC2 | stage IIIC2 | TNM Edition8 UICC | | | 0‑S | IV | stage IV | TNM Edition8 UICC | each T, each N, M1 | | 1‑L | IVA | stage IVA | TNM Edition8 UICC | organ dependent
| | 1‑L | IVB | stage IVB | TNM Edition8 UICC | organ dependent
| | 1‑L | IVC | stage IVC | TNM Edition8 UICC | organ dependent
| | 0‑L | BulkyIIA | stage Bulky IIA | TNM Edition8 UICC | | | 0‑L | BulkyIIB | stage Bulky IIB | TNM Edition8 UICC | | | 0‑L | ASKU | Asked, but unknown | Null Flavor | |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.7 UICC/AJCC T category 1.3.6.1.4.1.19376.1.3.11.43
| Id | 1.3.6.1.4.1.19376.1.3.11.43 | Effective Date | valid from 2017‑07‑07 15:06:39 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-t-value-8ed | Display Name | UICC T category |
|---|
| Description | T category in UICC/AJCC (TNM) 8th edition |
|---|
| 3 Source Code Systems | 2.16.840.1.113883.15.6 - TNM Edition7 UICC 2.16.840.1.113883.5.1008 - Null Flavor 2.16.840.1.113883.15.16 - TNM Edition8 UICC |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | Ta | Ta | TNM Edition7 UICC | | 0‑L | Tis | Tis | TNM Edition7 UICC | | 1‑L | Tis(DCIS) | Tis(DCIS) | TNM Edition7 UICC | | 1‑L | Tis(LCIS) | Tis(LCIS) | TNM Edition7 UICC | | 1‑L | Tis(Paget) | Tis(Paget) | TNM Edition7 UICC | | 1‑L | Tis(pu) | Tis(pu) | TNM Edition7 UICC | | 1‑L | Tis(pd) | Tis(pd) | TNM Edition7 UICC | | 0‑L | T0 | T0 | TNM Edition7 UICC | | 0‑L | T1 | T1 | TNM Edition7 UICC | | 1‑L | T1mi | T1mi | TNM Edition7 UICC | | 1‑L | T1a | T1a | TNM Edition7 UICC | | 2‑L | T1a1 | T1a1 | TNM Edition7 UICC | | 2‑L | T1a2 | T1a2 | TNM Edition7 UICC | | 1‑L | T1b | T1b | TNM Edition7 UICC | | 2‑L | T1b1 | T1b1 | TNM Edition7 UICC | | 2‑L | T1b2 | T1b2 | TNM Edition7 UICC | | 1‑L | T1c | T1c | TNM Edition7 UICC | | 1‑L | T1d | T1d | TNM Edition7 UICC | | 0‑L | T2 | T2 | TNM Edition7 UICC | | 1‑L | T2a | T2a | TNM Edition7 UICC | | 2‑L | T2a1 | T2a1 | TNM Edition7 UICC | | 2‑L | T2a2 | T2a2 | TNM Edition7 UICC | | 1‑L | T2b | T2b | TNM Edition7 UICC | | 1‑L | T2c | T2c | TNM Edition7 UICC | | 1‑L | T2d | T2d | TNM Edition7 UICC | | 0‑L | T3 | T3 | TNM Edition7 UICC | | 1‑L | T3a | T3a | TNM Edition7 UICC | | 1‑L | T3b | T3b | TNM Edition7 UICC | | 1‑L | T3c | T3c | TNM Edition7 UICC | | 1‑L | T3d | T3d | TNM Edition7 UICC | | 0‑L | T4 | T4 | TNM Edition7 UICC | | 1‑L | T4a | T4a | TNM Edition7 UICC | | 1‑L | T4b | T4b | TNM Edition7 UICC | | 1‑L | T4c | T4c | TNM Edition7 UICC | | 1‑L | T4d | T4d | TNM Edition7 UICC | | 1‑L | T4e | T4e | TNM Edition7 UICC | | 0‑L | Tx | TX | TNM Edition7 UICC | | 0‑L | Ta | Ta | TNM Edition8 UICC | | 0‑L | Tis | Tis | TNM Edition8 UICC | | 1‑L | Tis(LAMN) | Tis(LAMN) | TNM Edition8 UICC | | 1‑L | Tis(DCIS) | Tis(DCIS) | TNM Edition8 UICC | | 1‑L | Tis(LCIS) | Tis(LCIS) | TNM Edition8 UICC | | 1‑L | Tis(Paget) | Tis(Paget) | TNM Edition8 UICC | | 1‑L | Tis(pu) | Tis(pu) | TNM Edition8 UICC | | 1‑L | Tis(pd) | Tis(pd) | TNM Edition8 UICC | | 0‑L | T0 | T0 | TNM Edition8 UICC | | 0‑L | T1 | T1 | TNM Edition8 UICC | | 1‑L | T1mi | T1mi | TNM Edition8 UICC | | 1‑L | T1a | T1a | TNM Edition8 UICC | | 2‑L | T1a1 | T1a1 | TNM Edition8 UICC | | 2‑L | T1a2 | T1a2 | TNM Edition8 UICC | | 1‑L | T1b | T1b | TNM Edition8 UICC | | 2‑L | T1b1 | T1b1 | TNM Edition8 UICC | | 2‑L | T1b2 | T1b2 | TNM Edition8 UICC | | 1‑L | T1c | T1c | TNM Edition8 UICC | | 2‑L | T1c1 | T1c1 | TNM Edition8 UICC | | 2‑L | T1c2 | T1c2 | TNM Edition8 UICC | | 2‑L | T1c3 | T1c3 | TNM Edition8 UICC | | 1‑L | T1d | T1d | TNM Edition8 UICC | | 0‑L | T2 | T2 | TNM Edition8 UICC | | 1‑L | T2a | T2a | TNM Edition8 UICC | | 2‑L | T2a1 | T2a1 | TNM Edition8 UICC | | 2‑L | T2a2 | T2a2 | TNM Edition8 UICC | | 1‑L | T2b | T2b | TNM Edition8 UICC | | 1‑L | T2c | T2c | TNM Edition8 UICC | | 1‑L | T2d | T2d | TNM Edition8 UICC | | 0‑L | T3 | T3 | TNM Edition8 UICC | | 1‑L | T3a | T3a | TNM Edition8 UICC | | 1‑L | T3b | T3b | TNM Edition8 UICC | | 1‑L | T3c | T3c | TNM Edition8 UICC | | 1‑L | T3d | T3d | TNM Edition8 UICC | | 1‑L | T3e | T3e | TNM Edition8 UICC | | 0‑L | T4 | T4 | TNM Edition8 UICC | | 1‑L | T4a | T4a | TNM Edition8 UICC | | 1‑L | T4b | T4b | TNM Edition8 UICC | | 1‑L | T4c | T4c | TNM Edition8 UICC | | 1‑L | T4d | T4d | TNM Edition8 UICC | | 1‑L | T4e | T4e | TNM Edition8 UICC | | 0‑L | TX | TX | TNM Edition8 UICC |
| | 0‑L | ASKU | Asked, but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.8 UICC/AJCC T category (SCT) 1.3.6.1.4.1.19376.1.3.11.12
| Id | 1.3.6.1.4.1.19376.1.3.11.12 | Effective Date | valid from 2014‑05‑13 15:52:43 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-t-value5 | Display Name | UICC T category (SCT) |
|---|
| Description | T-Value from UICC classification |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 261663004 | Tumor stage Ta (tumor staging) | SNOMED Clinical Terms | | 0‑S | 44401000 | Tis category (finding) | SNOMED Clinical Terms | | 1‑L | 313145008 | Tumor stage Tis pu (finding) | SNOMED Clinical Terms | | 1‑L | 313146009 | Tumor stage Tis pd (finding) | SNOMED Clinical Terms | | 0‑L | 58790005 | T0 category (finding) | SNOMED Clinical Terms | | 0‑S | 23351008 | T1 category (finding) | SNOMED Clinical Terms | | 1‑L | 313108002 | Tumor stage T1mic (finding) | SNOMED Clinical Terms | | 1‑L | 261646003 | Tumor stage T1a (finding) | SNOMED Clinical Terms | | 2‑L | 261647007 | Tumor stage T1a1 (finding) | SNOMED Clinical Terms | | 2‑L | 261648002 | Tumor stage T1a2 (finding) | SNOMED Clinical Terms | | 1‑L | 261649005 | Tumor stage T1b (finding) | SNOMED Clinical Terms | | 2‑L | 313110000 | Tumor stage T1b1 (finding) | SNOMED Clinical Terms | | 2‑L | 313111001 | Tumor stage T1b2 (finding) | SNOMED Clinical Terms | | 1‑L | 261650005 | Tumor stage T1c (finding) | SNOMED Clinical Terms | | 0‑S | 67673008 | T2 category (finding) | SNOMED Clinical Terms | | 1‑L | 261651009 | Tumor stage T2a (finding) | SNOMED Clinical Terms | | 1‑L | 261652002 | Tumor stage T2b (finding) | SNOMED Clinical Terms | | 1‑L | 261653007 | Tumor stage T2c (finding) | SNOMED Clinical Terms | | 0‑S | 14410001 | T3 category (finding) | SNOMED Clinical Terms | | 1‑L | 261654001 | Tumor stage T3a (finding) | SNOMED Clinical Terms | | 1‑L | 261655000 | Tumor stage T3b (finding) | SNOMED Clinical Terms | | 2‑L | 261656004 | Tumor stage T3bi (finding) | SNOMED Clinical Terms | | 2‑L | 261657008 | Tumor stage T3bii (finding) | SNOMED Clinical Terms | | 1‑L | 261658003 | Tumor stage T3c (finding) | SNOMED Clinical Terms | | 0‑S | 65565005 | T4 category (finding) | SNOMED Clinical Terms | | 1‑L | 261659006 | Tumor stage T4a (finding) | SNOMED Clinical Terms | | 1‑L | 261660001 | Tumor stage T4b (finding) | SNOMED Clinical Terms | | 1‑L | 261661002 | Tumor stage T4c (finding) | SNOMED Clinical Terms | | 1‑L | 261662009 | Tumor stage T4d (finding) | SNOMED Clinical Terms | | 0‑L | 67101007 | TX category (finding) | SNOMED Clinical Terms |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.9 UICC/AJCC N category 1.3.6.1.4.1.19376.1.3.11.44
| Id | 1.3.6.1.4.1.19376.1.3.11.44 | Effective Date | valid from 2017‑07‑21 10:00:53 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC_n_value_8ed | Display Name | UICC N category |
|---|
| Description | UICC N category, 8th edition |
|---|
| 3 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.15.6 - TNM Edition7 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | N0 | N0 | TNM Edition8 UICC | | 0‑L | N1 | N1 | TNM Edition8 UICC | | 1‑L | N1a | N1a | TNM Edition8 UICC | | 1‑L | N1b | N1b | TNM Edition8 UICC | | 1‑L | N1c | N1c | TNM Edition8 UICC | | 0‑L | N2 | N2 | TNM Edition8 UICC | | 1‑L | N2a | N2a | TNM Edition8 UICC | | 1‑L | N2b | N2b | TNM Edition8 UICC | | 1‑L | N2c | N2c | TNM Edition8 UICC | | 0‑L | N3 | N3 | TNM Edition8 UICC | | 1‑L | N3a | N3a | TNM Edition8 UICC | | 1‑L | N3b | N3b | TNM Edition8 UICC | | 1‑L | N3c | N3c | TNM Edition8 UICC | | 0‑L | NX | Nx | TNM Edition8 UICC | | 0‑L | N0 | N0 | TNM Edition7 UICC | | 0‑L | N1 | N1 | TNM Edition7 UICC | | 1‑L | N1mi | N1mi | TNM Edition7 UICC | | 1‑L | N1a | N1a | TNM Edition7 UICC | | 1‑L | N1b | N1b | TNM Edition7 UICC | | 1‑L | N1c | N1c | TNM Edition7 UICC | | 0‑L | N2 | N2 | TNM Edition7 UICC | | 1‑L | N2mi | N2mi | TNM Edition7 UICC | | 1‑L | N2a | N2a | TNM Edition7 UICC | | 1‑L | N2b | N2b | TNM Edition7 UICC | | 1‑L | N2c | N2c | TNM Edition7 UICC | | 0‑L | N3 | N3 | TNM Edition7 UICC | | 1‑L | N3mi | N3mi | TNM Edition7 UICC | | 1‑L | N3a | N3a | TNM Edition7 UICC | | 1‑L | N3b | N3b | TNM Edition7 UICC | | 1‑L | N3c | N3c | TNM Edition7 UICC | | 0‑L | NX | Nx | TNM Edition7 UICC |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.10 UICC/AJCC N category (SCT) 1.3.6.1.4.1.19376.1.3.11.13
| Id | 1.3.6.1.4.1.19376.1.3.11.13 | Effective Date | valid from 2014‑10‑27 14:47:35 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC_N_values | Display Name | UICC N category (SCT) |
|---|
| Description | N-Value from UICC classification |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 62455006 | N0 category (finding) | SNOMED Clinical Terms | | 0‑L | 53623008 | N1 category (finding) | SNOMED Clinical Terms | | 1‑L | 277672002 | Node stage N1a (finding) | SNOMED Clinical Terms | | 1‑L | 277674001 | Node stage N1b (finding) | SNOMED Clinical Terms | | 2‑L | 277772008 | Node stage N1bi (finding) | SNOMED Clinical Terms | | 2‑L | 277776006 | Node stage N1bii (finding) | SNOMED Clinical Terms | | 2‑L | 277777002 | Node stage N1biii (finding) | SNOMED Clinical Terms | | 2‑L | 277778007 | Node stage N1biv (finding) | SNOMED Clinical Terms | | 1‑L | 369996002 | N1c: >7 nodes involved (finding) | SNOMED Clinical Terms | | 0‑L | 46059003 | N2 category (finding) | SNOMED Clinical Terms | | 1‑L | 261967001 | Node stage N2a (finding) | SNOMED Clinical Terms | | 1‑L | 261968006 | Node stage N2b (finding) | SNOMED Clinical Terms | | 1‑L | 261969003 | Node stage N2c (finding) | SNOMED Clinical Terms | | 0‑L | 5856006 | N3 category (finding) | SNOMED Clinical Terms | | 1‑L | 370012005 | N3a: > 6 cm in dimension (finding) | SNOMED Clinical Terms | | 1‑L | 370013000 | N3b: extension to supraclavicular fossa (finding) | SNOMED Clinical Terms | | 0‑L | 79420006 | NX category (finding) | SNOMED Clinical Terms |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.11 UICC/AJCC M category 1.3.6.1.4.1.19376.1.3.11.45
| Id | 1.3.6.1.4.1.19376.1.3.11.45 | Effective Date | valid from 2017‑07‑21 10:31:38 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-m-values-8ed | Display Name | UICC M category |
|---|
| 3 Source Code Systems | 2.16.840.1.113883.15.6 - TNM Edition7 UICC 2.16.840.1.113883.5.1008 - Null Flavor 2.16.840.1.113883.15.16 - TNM Edition8 UICC |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | M0 | M0 | TNM Edition7 UICC | | 0‑L | M1 | M1 | TNM Edition7 UICC | | 1‑L | M1mi | M1(mi) | TNM Edition7 UICC | | 1‑L | M1a | M1a | TNM Edition7 UICC | | 1‑L | M1b | M1b | TNM Edition7 UICC | | 1‑L | M1c | M1c | TNM Edition7 UICC | | 1‑L | M1d | M1d | TNM Edition7 UICC | | 1‑L | M1e | M1e | TNM Edition7 UICC | | 1‑L | M1cy | M1(cy) | TNM Edition7 UICC | | 0‑L | MX | MX | TNM Edition7 UICC | | 0‑L | M0 | M0 | TNM Edition8 UICC | | 0‑L | M1 | M1 | TNM Edition8 UICC | | 1‑L | M1a | M1a | TNM Edition8 UICC | | 1‑L | M1b | M1b | TNM Edition8 UICC | | 1‑L | M1c | M1c | TNM Edition8 UICC | | 1‑L | M1d | M1d | TNM Edition8 UICC | | 1‑L | M1cy | M1(cy) | TNM Edition8 UICC | | 0‑L | MX | MX | TNM Edition8 UICC |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.12 UICC/AJCC M category (SCT) 1.3.6.1.4.1.19376.1.3.11.14
| Id | 1.3.6.1.4.1.19376.1.3.11.14 | Effective Date | valid from 2014‑05‑13 16:11:15 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-m-value | Display Name | UICC M category (SCT) |
|---|
| Description | M-Value from UICC classification |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 30893008 | M0 category (finding) | SNOMED Clinical Terms | | 0‑L | 55440008 | M1 category (finding) | SNOMED Clinical Terms | | 1‑L | 261927002 | Metastasis stage M1a (finding) | SNOMED Clinical Terms | | 1‑L | 261928007 | Metastasis stage M1b (finding) | SNOMED Clinical Terms | | 1‑L | 261929004 | Metastasis stage M1c (finding) | SNOMED Clinical Terms | | 0‑L | 27167007 | MX category (finding) | SNOMED Clinical Terms |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.13 UICC/AJCC C-Factor 1.3.6.1.4.1.19376.1.3.11.19
This value set has values from TNM 7th edition only. There is no C-Factor in the 8th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.19 | Effective Date | valid from 2015‑11‑13 13:56:23 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | uicc-c-factor-tnm | Display Name | UICC C-Factor |
|---|
| Description | Qualifier for any TNM category coding the certainty of classification by the methods used
|
|---|
| Source Code System | 2.16.840.1.113883.15.6 - TNM Edition7 UICC |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | C1 | C1 | TNM Edition7 UICC | | 0‑L | C2 | C2 | TNM Edition7 UICC | | 0‑L | C3 | C3 | TNM Edition7 UICC | | 0‑L | C4 | C4 | TNM Edition7 UICC | | 0‑L | C5 | C5 | TNM Edition7 UICC |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.14 UICC/AJCC C-Factor (SCT) 1.3.6.1.4.1.19376.1.3.11.20
| Id | 1.3.6.1.4.1.19376.1.3.11.20 | Effective Date | valid from 2015‑06‑22 15:47:55 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | uicc-c-factor | Display Name | UICC C-Factor (SCT) |
|---|
| Description | Qualifier for any TNM category coding the certainty of classification by the methods used |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| Source Code System | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 84068000 | C1 - TNM certainty factor | SNOMED Clinical Terms | | 0‑L | 76321005 | C2 - TNM certainty factor | SNOMED Clinical Terms | | 0‑L | 44756004 | C3 - TNM certainty factor | SNOMED Clinical Terms | | 0‑L | 75800005 | C4 - TNM certainty factor | SNOMED Clinical Terms | | 0‑L | 59397002 | C5 - TNM certainty factor | SNOMED Clinical Terms |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.15 UICC/AJCC Residual tumor (R) classification 1.3.6.1.4.1.19376.1.3.11.21
| Id | 1.3.6.1.4.1.19376.1.3.11.21 | Effective Date | valid from 2015‑06‑25 11:23:51 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-R-status | Display Name | UICC Residual tumor (R) classification |
|---|
| Description | Value set for Residual tumor state with values for extended R-Klassifikation (TNM Suppl. 4th ed.) |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 5 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.15.6 - TNM Edition7 UICC 1.2.276.0.76.3.1.131.1.5.120 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | 258254000 | Residual tumor stage R0 (finding) | SNOMED Clinical Terms | | | 1‑L | 415675004 | Surgical circumferential margin is closest uninvolved margin to malignant neoplasm (finding) | SNOMED Clinical Terms | No residual tumor: R0 > 1mm CRM negative | | 0‑L | 278271003 | Residual tumor stage R1 (finding) | SNOMED Clinical Terms | | | 1‑L | 384620009 | Surgical circumferential margin involved by malignant neoplasm (tumor present 0-1 mm from circumferential resection margin) (finding) | SNOMED Clinical Terms | | | 0‑L | 278272005 | Residual tumor stage R2 (finding) | SNOMED Clinical Terms | | | 0‑L | 258253006 | Residual tumor stage RX (finding) | SNOMED Clinical Terms | | | 0‑L | RX | RX | TNM Edition8 UICC | | | 0‑L | R0>1mm | R0 > 1mm | TNM Edition7 UICC | | | 0‑L | R0<1mm | R0 < 1mm | TNM Edition7 UICC | | | 0‑L | R1 | R1 | TNM Edition8 UICC | | | 1‑L | R1-dir | R1-dir | TNM Edition7 UICC | | | 1‑L | R1(is) | R1(is) | 1.2.276.0.76.3.1.131.1.5.120 | | | 1‑L | R1(cy) | R1(cy) | 1.2.276.0.76.3.1.131.1.5.120 | | | 0‑L | R2 | R2 | TNM Edition8 UICC | | | 1‑L | R2a | R2a | TNM Edition7 UICC | | | 1‑L | R2b | R2b | TNM Edition7 UICC | | | 1‑L | R2c | R2c | TNM Edition7 UICC | | | 0‑L | CRM+ | CRM positive | TNM Edition7 UICC | | | 0‑L | CRM- | CRM negative | TNM Edition7 UICC | |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.16 UICC/AJCC clinical/pathological 1.3.6.1.4.1.19376.1.3.11.22
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.22 | Effective Date | valid from 2015‑11‑04 15:52:34 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-clinical-pathological | Display Name | UICC clinical/pathological |
|---|
| Description | Qualifiers for each TNM category classifying the observation method and the time frame of observation in relation to treatment |
|---|
| 3 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 1.2.276.0.76.11.9 - hl7de-valueset-9 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | p | p | TNM Edition8 UICC | The pathological assessment of the primary tumor entails a resection of the primary tumor or a biopsy adequate to evaluate the highest T category. The pathological assessment of the regional lymph nodes entails a removal of lymph nodes adequate to validate the absence of regional lymph node metastasis or sufficient to evaluate the highest pN category. The pathological assessment of distant metastasis entails microscopic examination. | | 0‑L | c | c | TNM Edition8 UICC | The clinical classification is based on evidence acquired before treatment | | 1‑L | u | u | hl7de-valueset-9 | The clinical classification is based on ultrasound evidence acquired before treatment | | 1‑L | ct | ct | hl7de-valueset-9 | The clinical classification is based on CT imaging evidence acquired before treatment | | 1‑L | mr | mr | hl7de-valueset-9 | The clinical classification is based on magnetic resonance imaging evidence acquired before treatment |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.17 UICC/AJCC Venous Invasion 1.3.6.1.4.1.19376.1.3.11.23
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.23 | Effective Date | valid from 2015‑11‑04 16:17:55 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-vene-invasion | Display Name | UICC Venous Invasion |
|---|
| Description | Observation for tumor spread coding the degree of venous invasion
|
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | V0 | V0 | TNM Edition8 UICC | | 0‑L | V1 | V1 | TNM Edition8 UICC | | 0‑L | V2 | V2 | TNM Edition8 UICC | | 0‑L | VX | VX | TNM Edition8 UICC |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.18 UICC/AJCC Lymphatic Invasion 1.3.6.1.4.1.19376.1.3.11.24
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.24 | Effective Date | valid from 2015‑11‑04 16:19:20 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-lymphatic-invasion | Display Name | UICC Lymphatic Invasion |
|---|
| Description | Observation for tumor spread coding the degree of lymphatic invasion |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | L0 | L0 | TNM Edition8 UICC | | 0‑L | L1 | L1 | TNM Edition8 UICC | | 0‑L | LX | LX | TNM Edition8 UICC |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.19 UICC/AJCC Perineurial Invasion 1.3.6.1.4.1.19376.1.3.11.25
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.25 | Effective Date | valid from 2015‑11‑04 16:20:42 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-perineural-invasion | Display Name | UICC Perineural Invasion |
|---|
| Description |
Observation for tumor spread coding the degree of perineural (lymphatic nerve sheet) invasion
|
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | Pn0 | Pn0 | TNM Edition8 UICC | no perineural invasion
| | 0‑L | Pn1 | Pn1 | TNM Edition8 UICC | Perineural invasion
| | 0‑L | PnX | PnX | TNM Edition8 UICC | unknown
|
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.20 UICC/AJCC Mucosa 1.3.6.1.4.1.19376.1.3.11.26
| Id | 1.3.6.1.4.1.19376.1.3.11.26 | Effective Date | valid from 2015‑11‑04 16:22:04 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-mucosa | Display Name | UICC Mucosa |
|---|
| Source Code System | 1.2.276.0.76.3.1.131.1.5.1 |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | m1 | m1 | 1.2.276.0.76.3.1.131.1.5.1 | restricted to mucosa, 1st third
| | 0‑L | m2 | m2 | 1.2.276.0.76.3.1.131.1.5.1 | restricted to mucosa, 2nd third
| | 0‑L | m3 | m3 | 1.2.276.0.76.3.1.131.1.5.1 | restricted to mucosa, 3rd third
|
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.21 UICC/AJCC Submucosa 1.3.6.1.4.1.19376.1.3.11.27
| Id | 1.3.6.1.4.1.19376.1.3.11.27 | Effective Date | valid from 2015‑11‑04 16:23:24 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-submucosa | Display Name | UICC Submucosa |
|---|
| Source Code System | 1.2.276.0.76.3.1.131.1.5.1 |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | sm1 | sm1 | 1.2.276.0.76.3.1.131.1.5.1 | restricted to submucosa, 1st third
| | 0‑L | sm2 | sm2 | 1.2.276.0.76.3.1.131.1.5.1 | restricted to submucosa, 2nd third
| | 0‑L | sm3 | sm3 | 1.2.276.0.76.3.1.131.1.5.1 | restricted to submucosa, 3rd third
|
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.22 UICC/AJCC Localization of distant metastases 1.3.6.1.4.1.19376.1.3.11.28
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.28 | Effective Date | valid from 2015‑11‑04 16:26:53 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-distant-metastasis-localization | Display Name | UICC Localization of distant metastases |
|---|
| Description | Qualifier for M category coding the localization of distant metastasis in case of M1 or M2 |
|---|
| Source Code System | 2.16.840.1.113883.15.16 - TNM Edition8 UICC |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | PUL | PUL | TNM Edition8 UICC | | 0‑L | OSS | OSS | TNM Edition8 UICC | | 0‑L | HEP | HEP | TNM Edition8 UICC | | 0‑L | BRA | BRA | TNM Edition8 UICC | | 0‑L | LYM | LYM | TNM Edition8 UICC | | 0‑L | OTH | OTH | TNM Edition8 UICC | | 0‑L | MAR | MAR | TNM Edition8 UICC | | 0‑L | PLE | PLE | TNM Edition8 UICC | | 0‑L | PER | PER | TNM Edition8 UICC | | 0‑L | ADR | ADR | TNM Edition8 UICC | | 0‑L | SKI | SKI | TNM Edition8 UICC |
|
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.23 UICC/AJCC Grade 3 tiered 1.3.6.1.4.1.19376.1.3.11.29
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.29 | Effective Date | valid from 2015‑11‑13 19:36:21 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICCgrade | Display Name | UICC Grade 3 tiered |
|---|
| Description | Coding of the grade of differentiation (malignancy) of tumors, coded by a four-, three-, or two-tiered grading system, often using the score of a special grading system for certain tumor types |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | 1 | G1 | TNM Edition8 UICC | grade 1, well (highly) differentiated | | 0‑L | 2 | G2 | TNM Edition8 UICC | grade 2, moderately differentiated | | 0‑L | 3 | G3 | TNM Edition8 UICC | grade 3, poorly differentiated | | 0‑L | X | GX | TNM Edition8 UICC | cannot be assessed |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.24 UICC/AJCC Grade 2 tiered 1.3.6.1.4.1.19376.1.3.11.30
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.30 | Effective Date | valid from 2015‑11‑13 19:43:09 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICCgrade-2 | Display Name | UICC Grade 2 tiered |
|---|
| Description | Coding of the grade of differentiation (malignancy) of tumors, coded by a two-tiered grading system, sometimes using the score of a special grading system for certain tumor types |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | L | lG | TNM Edition8 UICC | low grade, combining grade 1 and 2 (except urothelial tumors: only grade 1) | | 0‑L | H | hG | TNM Edition8 UICC | low grade, combining grade 3 and 4 (except urothelial tumors: grade 2 through 4) | | 0‑L | X | X | TNM Edition8 UICC | cannot be assessed |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.25 UICC/AJCC Grade 4 tiered 1.3.6.1.4.1.19376.1.3.11.31
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.31 | Effective Date | valid from 2015‑11‑13 19:46:53 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICCgrade-4 | Display Name | UICC Grade 4 tiered |
|---|
| Description | Coding of the grade of differentiation (malignancy) of tumors, coded by a four-tiered grading system, sometimes using the score of a special grading system for certain tumor types. |
|---|
| 3 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 1.2.276.0.76.5.336 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | 1 | G1 | TNM Edition8 UICC | well (highly) differentiated | | 0‑L | 2 | G2 | TNM Edition8 UICC | moderately differentiated | | 0‑L | 3 | G3 | TNM Edition8 UICC | poorly differentiated | | 0‑L | 4 | G4 | TNM Edition8 UICC | undifferentiated resp. anaplastic | | 0‑L | X | GX | TNM Edition8 UICC | cannot be assessed | | 0‑L | 0 | 0 | 1.2.276.0.76.5.336 | Malignes Melanom der Konjunktiva | | 0‑L | X | X | 1.2.276.0.76.5.336 | nicht bestimmbar | | 0‑L | L | lG | 1.2.276.0.76.5.336 | Low grade (G1 oder G2) | | 0‑L | M | mG | 1.2.276.0.76.5.336 | intermediate (G2 oder G3) | | 0‑L | H | hG | 1.2.276.0.76.5.336 | high grade (G3 oder G4) | | 0‑L | B | B | 1.2.276.0.76.5.336 | Borderline-Malignität | | 0‑L | U | U | 1.2.276.0.76.5.336 | unbekannt | | 0‑L | T | T | 1.2.276.0.76.5.336 | trifft nicht zu |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.26 UICC/AJCC Tumor Serum Markers 1.3.6.1.4.1.19376.1.3.11.32
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.32 | Effective Date | valid from 2015‑11‑08 18:26:39 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICCSerumTumorMarkers | Display Name | UICC Serum Tumor Markers |
|---|
| Description | Observation of serum levels of tumor markers coded for germ cell tumors |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | S0 | S0 | TNM Edition8 UICC | serum tumor markers within normal limits | | 0‑L | S1 | S1 | TNM Edition8 UICC | LDH < 1,5* normal limit AND ßHCG< 5000mIU/ml AND AFP <1000 ng/ml
| | 0‑L | S2 | S2 | TNM Edition8 UICC | LDH 1,5 - 10* normal limit OR ßHCG 5000 - 50000 mIU/ml OR AFP 1000 - 10000 ng/ml
| | 0‑L | S3 | S3 | TNM Edition8 UICC | LDH >10* normal limit OR ßHCG > 50000 mIU/ml OR AFP > 10000 ng/ml
| | 0‑L | SX | SX | TNM Edition8 UICC | No information about serum tumor markers |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.27 ICD-O-3 Morphology and Behavior 1.3.6.1.4.1.19376.1.3.11.33
| Id | 1.3.6.1.4.1.19376.1.3.11.33 | Effective Date | valid from 2015‑11‑13 14:08:46 |
|---|
| Status |  Draft Draft | Version Label | 1st rev. (2013) |
|---|
| Name | icd-o-3-m | Display Name | ICD-O-3 Morphology |
|---|
| Description | International classification of diseases / oncology, morphological part for typing of benign and malignant tumors. The morphology axis provides six-digit codes ranging from 8000/0 to 9989/3. The first four digits indicate the specific histological term, i.e. morphology (8000 through 9992). <span style="text-align: justify;">In developing the previous editions and the present edition of ICD-O, a particular effort was made to use the nomenclature appearing in the World Health Organization </span>
<span style="text-align: justify;">International Histological Classification of Tumours</span> <span style="text-align: justify;">series (WHO “Blue Books”)</span>
<span style="text-align: justify;">7</span>
<span style="text-align: justify;">. This series covers all the principal sites of cancer and includes the morphology codes of ICD-O for each neoplasm. </span>All new histological classifications or modifications have been made by the WHO / IARC in so-called "Blue Books" by  2011 are listed on-line: http://codes.iarc.fr/changes.php.<br clear="none"/>The fifth digit after a slash (/) is the behavior code, which indicates whether a tumor is malignant, benign, in situ, or uncertain (whether benign or malignant). <br clear="none"/>A separate, 6th, one-digit code may also be given for histologic 4-tiered grading (differentiation) or for the cell type in lymphatic neoplasia.  <br clear="none"/>The fifth and the sixth digit are coded separately in value sets 1.3.6.1.4.1.19376.1.3.11.34 and 1.3.6.1.4.1.19376.1.3.11.35 |
|---|
A valid code from the code system:| Code System Name | Code System Id | Code System Version | | International Classification of Diseases for Oncology, version 3. | 2.16.840.1.113883.6.43.1 | |
|
| Or one of the following: |
| 2 Source Code Systems | 2.16.840.1.113883.6.43.1 - International Classification of Diseases for Oncology, version 3. 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | | Code System |
|---|
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.28 ICD-O-3 Behavior 1.3.6.1.4.1.19376.1.3.11.34
| Id | 1.3.6.1.4.1.19376.1.3.11.34 | Effective Date | valid from 2015‑11‑13 18:54:30 |
|---|
| Status |  Draft Draft | Version Label | 1st rev. (2013) |
|---|
| Name | icd-o-3-behavior | Display Name | ICD-O-3 Behavior |
|---|
| Description | Coding of the behavior of a tumor, coded by the first digit after slash in ICD-O-Morphology code |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.43.1 - International Classification of Diseases for Oncology, version 3. 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | 0 | /0 | International Classification of Diseases for Oncology, version 3. | benign | | 0‑L | 1 | /1 | International Classification of Diseases for Oncology, version 3. | borderline malignancy or uncertain malignant potential | | 0‑L | 2 | /2 | International Classification of Diseases for Oncology, version 3. | noninvasiv resp. carcinoma in situ | | 0‑L | 3 | /3 | International Classification of Diseases for Oncology, version 3. | invasive resp. malignant primary tumor | | 0‑L | 6 | /6 | International Classification of Diseases for Oncology, version 3. | metastasis resp. secondary tumor, not used in German cancer registries | | 0‑L | 9 | /9 | International Classification of Diseases for Oncology, version 3. | malignant, uncertain whether primary or secondary tumor, not used in German cancer registries |
| | 0‑L | ASKU | asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.29 ICD-O-3 Differentiation 1.3.6.1.4.1.19376.1.3.11.35
| Id | 1.3.6.1.4.1.19376.1.3.11.35 | Effective Date | valid from 2015‑11‑13 19:07:32 |
|---|
| Status |  Draft Draft | Version Label | 1st rev. (2013) |
|---|
| Name | icd-o-3-differentiation | Display Name | ICD-O-3 Differentiation |
|---|
| Description | Coding the cellular differentiation of malignant tumors, either the grade of malignancy (differentiation), or the cell line differentiation, coded by the second digit after the slash in ICD-O-3 Morphology axi |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.43.1 - International Classification of Diseases for Oncology, version 3. 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | 1 | 1 | International Classification of Diseases for Oncology, version 3. | well (highly) differentiated | | 0‑L | 2 | 2 | International Classification of Diseases for Oncology, version 3. | moderately differentiated | | 0‑L | 3 | 3 | International Classification of Diseases for Oncology, version 3. | poorly differentiated | | 0‑L | 4 | 4 | International Classification of Diseases for Oncology, version 3. | undifferentiated resp. anaplastic | | 0‑L | 5 | 5 | International Classification of Diseases for Oncology, version 3. | T-cell differentiation in lymphomas and leukemias | | 0‑L | 6 | 6 | International Classification of Diseases for Oncology, version 3. | B-cell differentiation, pre-B-cells, B-precursor cells in lymphomas and leukemias | | 0‑L | 7 | 7 | International Classification of Diseases for Oncology, version 3. | Null-cell, non-B-nonT-cell differentiation in lymphomas and leukemias | | 0‑L | 8 | 8 | International Classification of Diseases for Oncology, version 3. | NK (natural killer)-cell differentiation in lymphomas and leukemias | | 0‑L | 9 | 9 | International Classification of Diseases for Oncology, version 3. | grading not applied, not applicable, unknown; cell type not assessed or unknown |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.30 ICD-O-3 Topography 1.3.6.1.4.1.19376.1.3.11.36
| Id | 1.3.6.1.4.1.19376.1.3.11.36 | Effective Date | valid from 2015‑11‑13 19:16:23 |
|---|
| Status |  Draft Draft | Version Label | 1st rev. (2013) |
|---|
| Name | icd-o-3-topography | Display Name | ICD-O-3 Topography |
|---|
| Description | International classification of diseases / oncology, topographical part for typing of benign and malignant tumors. The topography axis (C-axis) uses the ICD-10 classification of malignant neoplasms (except those categories which relate to secondary neoplasms and to specified morphological types of tumours) for all types of tumours, thereby providing greater site detail for non-malignant tumours than is provided in ICD-10. In contrast to ICD-10, the ICD-O includes topography for sites of haematopoietic and reticuloendothelial tumors. |
|---|
A valid code from the code system:| Code System Name | Code System Id | Code System Version | | International Classification of Diseases for Oncology, version 3. | 2.16.840.1.113883.6.43.1 | |
|
| Or one of the following: |
| 2 Source Code Systems | 2.16.840.1.113883.6.43.1 - International Classification of Diseases for Oncology, version 3. 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | | Code System |
|---|
| | 0‑L | ASKU | asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.31 UICC/AJCC Multiplicity 1.3.6.1.4.1.19376.1.3.11.37
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.37 | Effective Date | valid from 2015‑11‑06 10:44:54 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-Multiplicity | Display Name | UICC Multiplicity |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | m | (m) | TNM Edition8 UICC | Qualifier for T-category: multiple simultaneous tumors in one organ | | 0‑L | 2 | (2) | TNM Edition8 UICC | Qualifier for T-category: two simultaneous tumors in one organ | | 0‑L | 3 | (3) | TNM Edition8 UICC | Qualifier for T-category: three simultaneous tumors in one organ | | 0‑L | 4 | (4) | TNM Edition8 UICC | Qualifier for T-category: four simultaneous tumors in one organ | | 0‑L | 5 | (5) | TNM Edition8 UICC | Qualifier for T-category: five simultaneous tumors in one organ | | 0‑L | 6 | (6) | TNM Edition8 UICC | Qualifier for T-category: six simultaneous tumors in one organ | | 0‑L | 7 | (7) | TNM Edition8 UICC | Qualifier for T-category: seven simultaneous tumors in one organ | | 0‑L | 8 | (8) | TNM Edition8 UICC | Qualifier for T-category: eight simultaneous tumors in one organ | | 0‑L | 9 | (9) | TNM Edition8 UICC | Qualifier for T-category: nine simultaneous tumors in one organ<br clear="none"/> | | 0‑L | 10 | (10) | TNM Edition8 UICC | Qualifier for T-category: ten simultaneous tumors in one organ<br clear="none"/> |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.32 UICC/AJCC Isolated Tumor Cells 1.3.6.1.4.1.19376.1.3.11.38
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.38 | Effective Date | valid from 2017‑03‑13 09:48:35 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICCITC | Display Name | UICC Isolated Tumor Cells |
|---|
| Description | Codes for isolated tumor cells, detected by immunohistochemical or molecular pathology methods in regional and non-regional lymph nodes, used in qualifiers for N- and M-category in TNM |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | i+ | (i+) | TNM Edition8 UICC | | 0‑L | i- | (i-) | TNM Edition8 UICC | | 0‑L | mol+ | (mol+) | TNM Edition8 UICC | | 0‑L | mol- | (mol-) | TNM Edition8 UICC |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.33 UICC/AJCC Recurrence 1.3.6.1.4.1.19376.1.3.11.39
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.39 | Effective Date | valid from 2015‑11‑06 10:36:10 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-Relapse | Display Name | UICC Recurrence |
|---|
| Description | Symbol for TNM: recurrent tumor after a disease-free interval after a curative treatment
|
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | r | r | TNM Edition8 UICC | Symbol for TNM: recurrent tumor after a disease-free interval after a curative treatment |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.34 UICC/AJCC Posttherapeutic 1.3.6.1.4.1.19376.1.3.11.40
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.40 | Effective Date | valid from 2015‑11‑06 10:39:53 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-Posttherapeutic | Display Name | UICC Posttherapeutic |
|---|
| Description | Symbol for TNM: tumor after a multimodal (e.g. neo-adjuvant) therapy
|
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | y | y | TNM Edition8 UICC | Symbol for TNM: tumor after a multimodal (e.g. neo-adjuvant) therapy
|
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.35 UICC/AJCC Autopsy 1.3.6.1.4.1.19376.1.3.11.41
This value set has values from TNM 8th edition. There is no difference to 7th edition.
| Id | 1.3.6.1.4.1.19376.1.3.11.41 | Effective Date | valid from 2015‑11‑18 15:55:17 |
|---|
| Status |  Draft Draft | Version Label | |
|---|
| Name | UICC-Autopsy | Display Name | UICC Autopsy |
|---|
| Description | Symbol for TNM: Has this tumor formula been derived first at autopsy? |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.15.16 - TNM Edition8 UICC 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System | Description |
|---|
| 0‑L | a | a | TNM Edition8 UICC | Symbol for TNM: Has this tumor formula been derived first at autopsy? |
| | 0‑L | ASKU | Asked but unknown | Null Flavor | |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
A.37 Clinical Significance 1.3.6.1.4.1.19376.1.3.11.47
| Id | 1.3.6.1.4.1.19376.1.3.11.47 | Effective Date | valid from 2017‑11‑15 11:08:46 |
|---|
| Status |  Draft Draft | Version Label | 2.0 |
|---|
| Name | ClinicalSignificance | Display Name | Clinical Significance |
|---|
| Description | Value set for expressing different degrees of significance for clinical decisions in case a pathology report has been updated |
|---|
| Copyright | This artefact includes content from SNOMED Clinical Terms® (SNOMED CT®) which is copyright of the International Health Terminology Standards Development Organisation (IHTSDO). Implementers of these artefacts must have the appropriate SNOMED CT Affiliate license - for more information contact http://www.snomed.org/snomed-ct/getsnomed-ct or info@snomed.org. |
|---|
| 2 Source Code Systems | 2.16.840.1.113883.6.96 - SNOMED Clinical Terms 2.16.840.1.113883.5.1008 - Null Flavor |
|---|
| Level/ Type | Code | Display Name | Code System |
|---|
| 0‑L | 371928007 | Not significant (qualifier value) | SNOMED Clinical Terms | | 0‑L | 371925005 | Most significant (qualifier value) | SNOMED Clinical Terms | | 0‑L | 371927002 | Moderately significant (qualifier value) | SNOMED Clinical Terms | | 0‑L | 386134007 | Significant (qualifier value) | SNOMED Clinical Terms | | 0‑L | 386135008 | Significance undetermined (qualifier value) | SNOMED Clinical Terms | | 0‑L | 371926006 | Highly significant (qualifier value) | SNOMED Clinical Terms |
| | 0‑L | ASKU | Asked but unknown | Null Flavor |
|
| Legenda: Type L=leaf, S=specializable, A=abstract, D=deprecated. NullFlavors to appear in @nullFlavor attribute instead of @code. |
References








